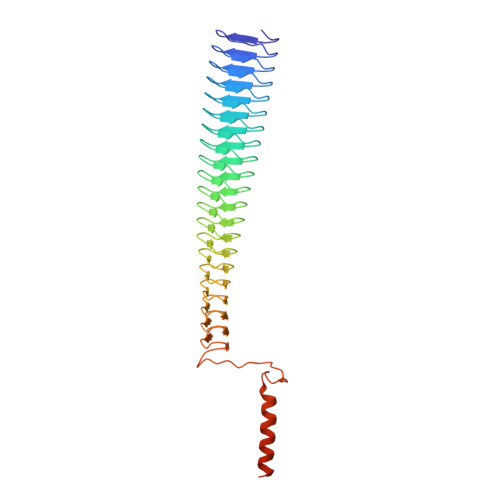
The C-terminal head domain of Burkholderia pseudomallei BpaC has a striking hydrophilic core with an extensive solvent network.
Kiessling, A.R., Harris, S.A., Weimer, K.M., Wells, G., Goldman, A.(2022) Mol Microbiol 118: 77-91
- PubMed: 35703459
- DOI: https://doi.org/10.1111/mmi.14953
- Primary Citation of Related Structures:
7O23 - PubMed Abstract:
Gram-negative pathogens like Burkholderia pseudomallei use trimeric autotransporter adhesins such as BpaC as key molecules in their pathogenicity. Our 1.4 Å crystal structure of the membrane-proximal part of the BpaC head domain shows that the domain is exclusively made of left-handed parallel β-roll repeats. This, the largest such structure solved, has two unique features. First, the core, rather than being composed of the canonical hydrophobic Ile and Val, is made up primarily of the hydrophilic Thr and Asn, with two different solvent channels. Second, comparing BpaC to all other left-handed parallel β-roll structures showed that the position of the head domain in the protein correlates with the number and type of charged residues. In BpaC, only negatively charged residues face the solvent-in stark contrast to the primarily positive surface charge of the left-handed parallel β-roll "type" protein, YadA. We propose extending the definitions of these head domains to include the BpaC-like head domain as a separate subtype, based on its unusual sequence, position, and charge. We speculate that the function of left-handed parallel β-roll structures may differ depending on their position in the structure.
Organizational Affiliation:
Astbury Centre for Structural Molecular Biology, School of Biomedical Sciences, University of Leeds, Leeds, UK.