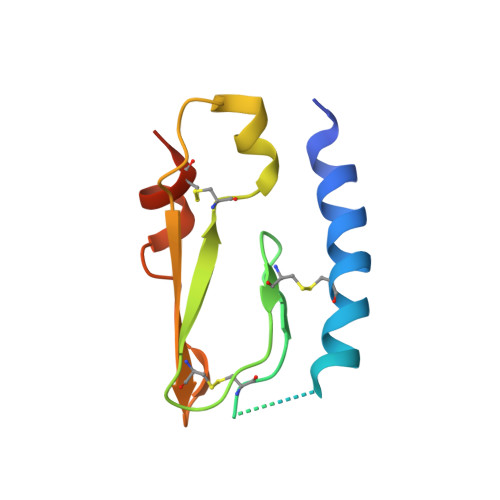
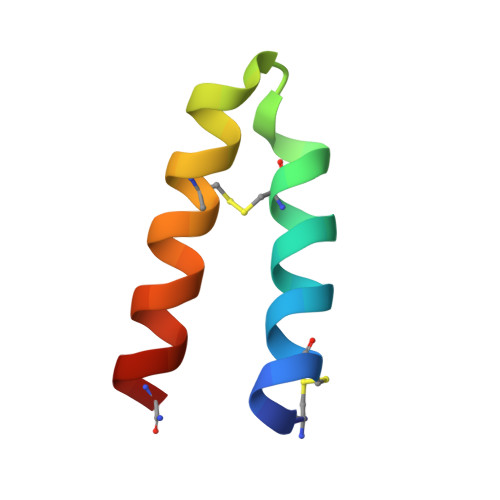
Discovery of Selective Pituitary Adenylate Cyclase 1 Receptor (PAC1R) Antagonist Peptides Potent in a Maxadilan/PACAP38-Induced Increase in Blood Flow Pharmacodynamic Model.
Hu, E., Hong, F.T., Aral, J., Long, J., Piper, D.E., Poppe, L., Andrews, K.L., Hager, T., Davis, C., Li, H., Wong, P., Gavva, N., Shi, L., Zhu, D.X.D., Lehto, S.G., Xu, C., Miranda, L.P.(2021) J Med Chem 64: 3427-3438
- PubMed: 33715378
- DOI: https://doi.org/10.1021/acs.jmedchem.0c01396
- Primary Citation of Related Structures:
7JQD - PubMed Abstract:
Inhibition of the pituitary adenylate cyclase 1 receptor (PAC1R) is a novel mechanism that could be used for abortive treatment of acute migraine. Our research began with comparative analysis of known PAC1R ligand scaffolds, PACAP38 and Maxadilan, which resulted in the selection of des(24-42) Maxadilan, 6 , as a starting point. C-terminal modifications of 6 improved the peptide metabolic stability in vitro and in vivo . SAR investigations identified synergistic combinations of amino acid replacements that significantly increased the in vitro PAC1R inhibitory activity of the analogs to the pM IC 90 range. Our modifications further enabled deletion of up to six residues without impacting potency, thus improving peptide ligand binding efficiency. Analogs 17 and 18 exhibited robust in vivo efficacy in the rat Maxadilan-induced increase in blood flow (MIIBF) pharmacodynamic model at 0.3 mg/kg subcutaneous dosing. The first cocrystal structure of a PAC1R antagonist peptide ( 18 ) with PAC1R extracellular domain is reported.
Organizational Affiliation:
Therapeutic Discovery, Amgen Inc., One Amgen Center Drive, Thousand Oaks, California 91320, United States.