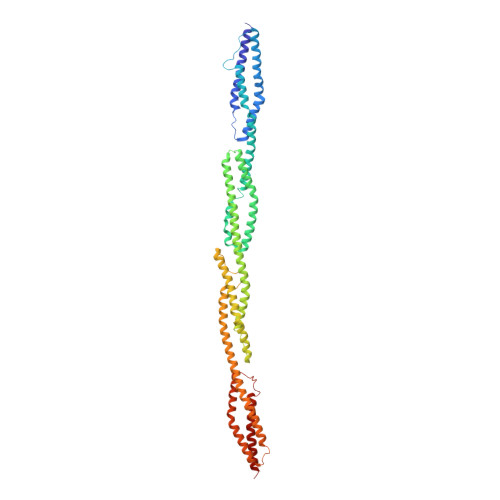Order from disorder in the sarcomere: FATZ forms a fuzzy but tight complex and phase-separated condensates with alpha-actinin.
Sponga, A., Arolas, J.L., Schwarz, T.C., Jeffries, C.M., Rodriguez Chamorro, A., Kostan, J., Ghisleni, A., Drepper, F., Polyansky, A., De Almeida Ribeiro, E., Pedron, M., Zawadzka-Kazimierczuk, A., Mlynek, G., Peterbauer, T., Doto, P., Schreiner, C., Hollerl, E., Mateos, B., Geist, L., Faulkner, G., Kozminski, W., Svergun, D.I., Warscheid, B., Zagrovic, B., Gautel, M., Konrat, R., Djinovic-Carugo, K.(2021) Sci Adv 7
- PubMed: 34049882
- DOI: https://doi.org/10.1126/sciadv.abg7653
- Primary Citation of Related Structures:
7A8T, 7A8U, 7ANK - PubMed Abstract:
In sarcomeres, α-actinin cross-links actin filaments and anchors them to the Z-disk. FATZ (filamin-, α-actinin-, and telethonin-binding protein of the Z-disk) proteins interact with α-actinin and other core Z-disk proteins, contributing to myofibril assembly and maintenance. Here, we report the first structure and its cellular validation of α-actinin-2 in complex with a Z-disk partner, FATZ-1, which is best described as a conformational ensemble. We show that FATZ-1 forms a tight fuzzy complex with α-actinin-2 and propose an interaction mechanism via main molecular recognition elements and secondary binding sites. The obtained integrative model reveals a polar architecture of the complex which, in combination with FATZ-1 multivalent scaffold function, might organize interaction partners and stabilize α-actinin-2 preferential orientation in Z-disk. Last, we uncover FATZ-1 ability to phase-separate and form biomolecular condensates with α-actinin-2, raising the question whether FATZ proteins can create an interaction hub for Z-disk proteins through membraneless compartmentalization during myofibrillogenesis.
Organizational Affiliation:
Department of Structural and Computational Biology, Max Perutz Labs, University of Vienna, Campus Vienna Biocenter 5, A-1030 Vienna, Austria.