The making of a potent L-lactate transport inhibitor
Bosshart, P.D., Kalbermatter, D., Bonetti, S., Fotiadis, D.(2021) Commun Chem 4: 128
Experimental Data Snapshot
(2021) Commun Chem 4: 128
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| L-lactate transporter | 420 | Syntrophobacter fumaroxidans MPOB | Mutation(s): 0 Gene Names: Sfum_3364 Membrane Entity: Yes | 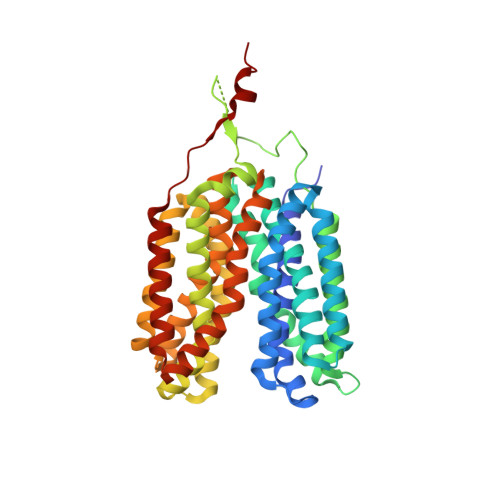 | |
UniProt | |||||
Find proteins for A0LNN5 (Syntrophobacter fumaroxidans (strain DSM 10017 / MPOB)) Explore A0LNN5 Go to UniProtKB: A0LNN5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0LNN5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 02Q (Subject of Investigation/LOI) Query on 02Q | C [auth A], D [auth B] | 3-(2-methylphenyl)propanoic acid C10 H12 O2 JIRKNEAMPYVPTD-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 102.048 | α = 90 |
| b = 199.703 | β = 90 |
| c = 61.616 | γ = 90 |
| Software Name | Purpose |
|---|---|
| Aimless | data scaling |
| PHASER | phasing |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Swiss National Science Foundation | Switzerland | 310030_184980 |