Discovery and Optimization of Novel SUCNR1 Inhibitors: Design of Zwitterionic Derivatives with a Salt Bridge for the Improvement of Oral Exposure.
Velcicky, J., Wilcken, R., Cotesta, S., Janser, P., Schlapbach, A., Wagner, T., Piechon, P., Villard, F., Bouhelal, R., Piller, F., Harlfinger, S., Stringer, R., Fehlmann, D., Kaupmann, K., Littlewood-Evans, A., Haffke, M., Gommermann, N.(2020) J Med Chem 63: 9856-9875
- PubMed: 32856916
- DOI: https://doi.org/10.1021/acs.jmedchem.0c01020
- Primary Citation of Related Structures:
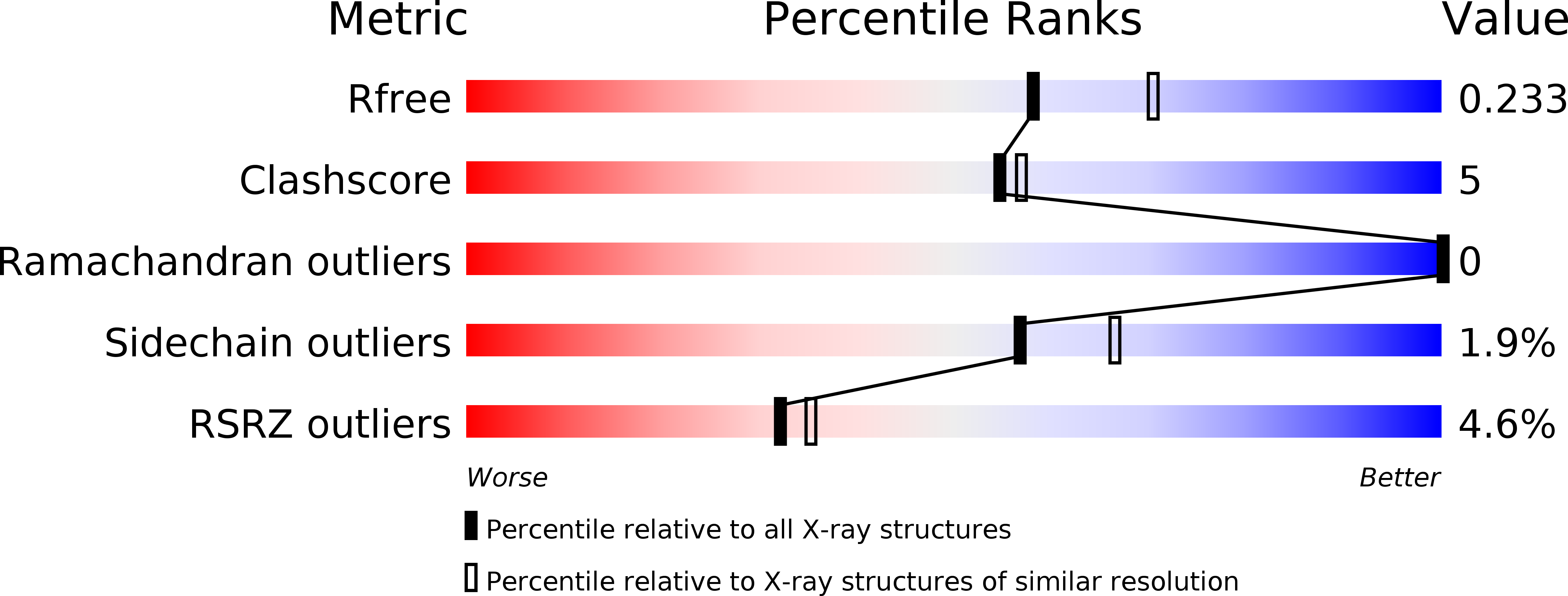
6Z10 - PubMed Abstract:
G-protein-coupled receptor SUCNR1 (succinate receptor 1 or GPR91) senses the citric cycle intermediate succinate and is implicated in various pathological conditions such as rheumatoid arthritis, liver fibrosis, or obesity. Here, we describe a novel SUCNR1 antagonist scaffold discovered by high-throughput screening. The poor permeation and absorption properties of the most potent compounds, which were zwitterionic in nature, could be improved by the formation of an internal salt bridge, which helped in shielding the two opposite charges and thus also the high polarity of zwitterions with separated charges. The designed compounds containing such a salt bridge reached high oral bioavailability and oral exposure. We believe that this principle could find a broad interest in the medicinal chemistry field as it can be useful not only for the modulation of properties in zwitterionic compounds but also in acidic or basic compounds with poor permeation.
Organizational Affiliation:
Novartis Institutes for BioMedical Research, CH-4002 Basel, Switzerland.