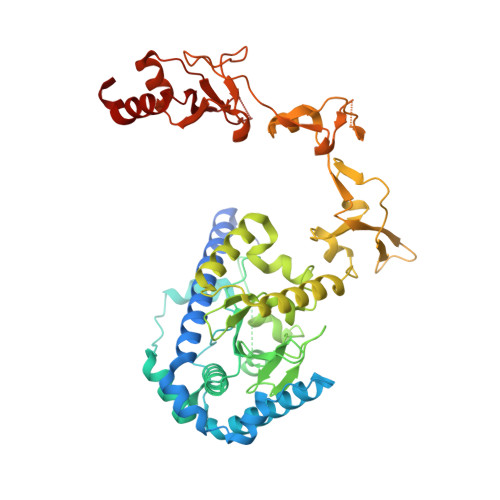
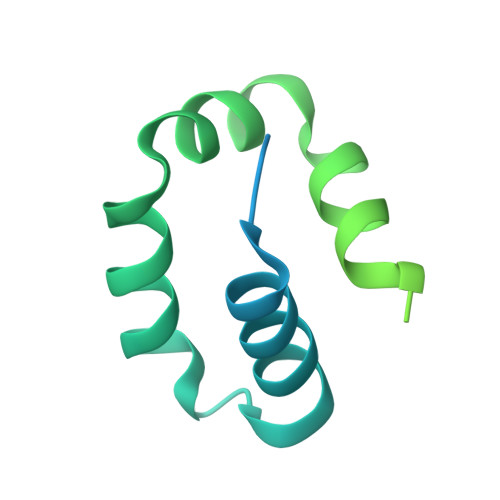
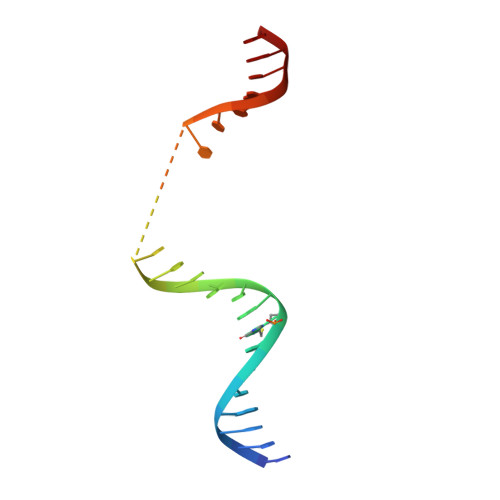
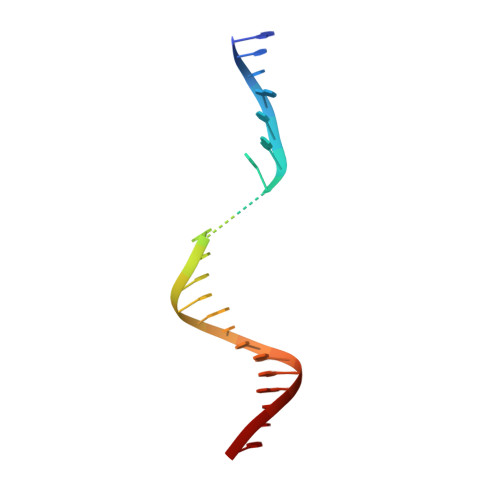
Tethering-facilitated DNA 'opening' and complementary roles of beta-hairpin motifs in the Rad4/XPC DNA damage sensor protein
Paul, D., Mu, H., Tavakoli, A., Dai, Q., Chen, X., Chakraborty, S., He, C., Ansari, A., Broyde, S., Min, J.H.(2021) Nucleic Acids Res 48: 12348-12364