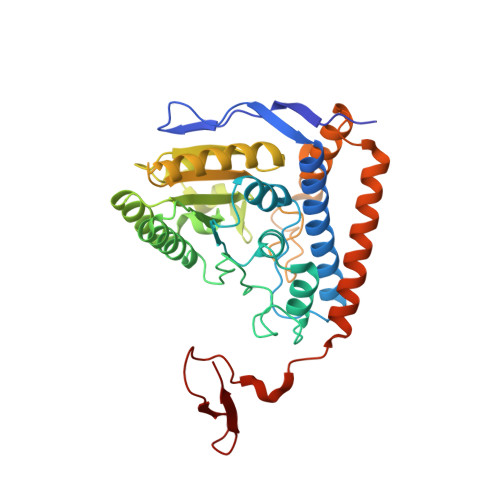
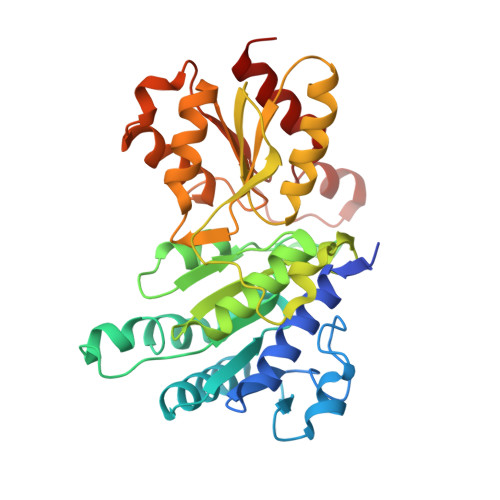
Pyruvate dehydrogenase complex deficiency is linked to regulatory loop disorder in the alpha V138M variant of human pyruvate dehydrogenase.
Whitley, M.J., Arjunan, P., Nemeria, N.S., Korotchkina, L.G., Park, Y.H., Patel, M.S., Jordan, F., Furey, W.(2018) J Biol Chem 293: 13204-13213
- PubMed: 29970614
- DOI: https://doi.org/10.1074/jbc.RA118.003996
- Primary Citation of Related Structures:
6CER, 6CFO - PubMed Abstract:
The pyruvate dehydrogenase multienzyme complex (PDHc) connects glycolysis to the tricarboxylic acid cycle by producing acetyl-CoA via the decarboxylation of pyruvate. Because of its pivotal role in glucose metabolism, this complex is closely regulated in mammals by reversible phosphorylation, the modulation of which is of interest in treating cancer, diabetes, and obesity. Mutations such as that leading to the αV138M variant in pyruvate dehydrogenase, the pyruvate-decarboxylating PDHc E1 component, can result in PDHc deficiency, an inborn error of metabolism that results in an array of symptoms such as lactic acidosis, progressive cognitive and neuromuscular deficits, and even death in infancy or childhood. Here we present an analysis of two X-ray crystal structures at 2.7-Å resolution, the first of the disease-associated human αV138M E1 variant and the second of human wildtype (WT) E1 with a bound adduct of its coenzyme thiamin diphosphate and the substrate analogue acetylphosphinate. The structures provide support for the role of regulatory loop disorder in E1 inactivation, and the αV138M variant structure also reveals that altered coenzyme binding can result in such disorder even in the absence of phosphorylation. Specifically, both E1 phosphorylation at αSer-264 and the αV138M substitution result in disordered loops that are not optimally oriented or available to efficiently bind the lipoyl domain of PDHc E2. Combined with an analysis of αV138M activity, these results underscore the general connection between regulatory loop disorder and loss of E1 catalytic efficiency.
Organizational Affiliation:
From the Department of Pharmacology and Chemical Biology, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania 15261.