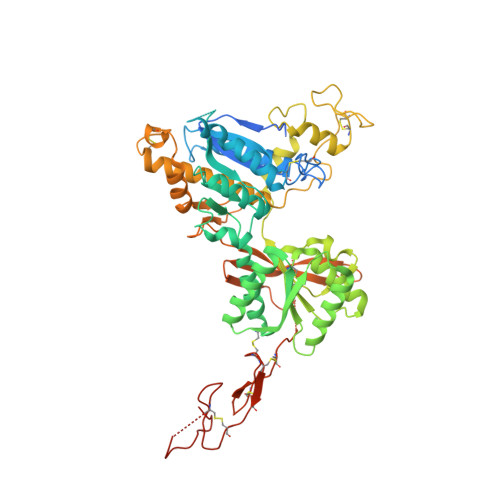
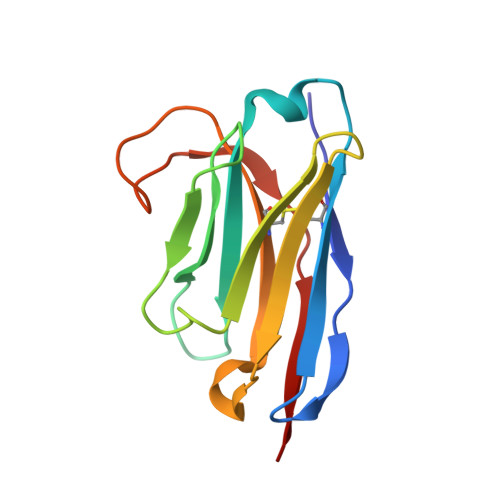
Structural insights into the activation of metabotropic glutamate receptors.
Koehl, A., Hu, H., Feng, D., Sun, B., Zhang, Y., Robertson, M.J., Chu, M., Kobilka, T.S., Laermans, T., Steyaert, J., Tarrasch, J., Dutta, S., Fonseca, R., Weis, W.I., Mathiesen, J.M., Skiniotis, G., Kobilka, B.K.(2019) Nature 566: 79-84
- PubMed: 30675062
- DOI: https://doi.org/10.1038/s41586-019-0881-4
- Primary Citation of Related Structures:
6N4X, 6N4Y, 6N50, 6N51, 6N52 - PubMed Abstract:
Metabotropic glutamate receptors are family C G-protein-coupled receptors. They form obligate dimers and possess extracellular ligand-binding Venus flytrap domains, which are linked by cysteine-rich domains to their 7-transmembrane domains. Spectroscopic studies show that signalling is a dynamic process, in which large-scale conformational changes underlie the transmission of signals from the extracellular Venus flytraps to the G protein-coupling domains-the 7-transmembrane domains-in the membrane. Here, using a combination of X-ray crystallography, cryo-electron microscopy and signalling studies, we present a structural framework for the activation mechanism of metabotropic glutamate receptor subtype 5. Our results show that agonist binding at the Venus flytraps leads to a compaction of the intersubunit dimer interface, thereby bringing the cysteine-rich domains into close proximity. Interactions between the cysteine-rich domains and the second extracellular loops of the receptor enable the rigid-body repositioning of the 7-transmembrane domains, which come into contact with each other to initiate signalling.
Organizational Affiliation:
Department of Structural Biology, Stanford University School of Medicine, Stanford, CA, USA.