A conserved CAF40-binding motif in metazoan NOT4 mediates association with the CCR4-NOT complex.
Keskeny, C., Raisch, T., Sgromo, A., Igreja, C., Bhandari, D., Weichenrieder, O., Izaurralde, E.(2019) Genes Dev 33: 236-252
- PubMed: 30692204
- DOI: https://doi.org/10.1101/gad.320952.118
- Primary Citation of Related Structures:
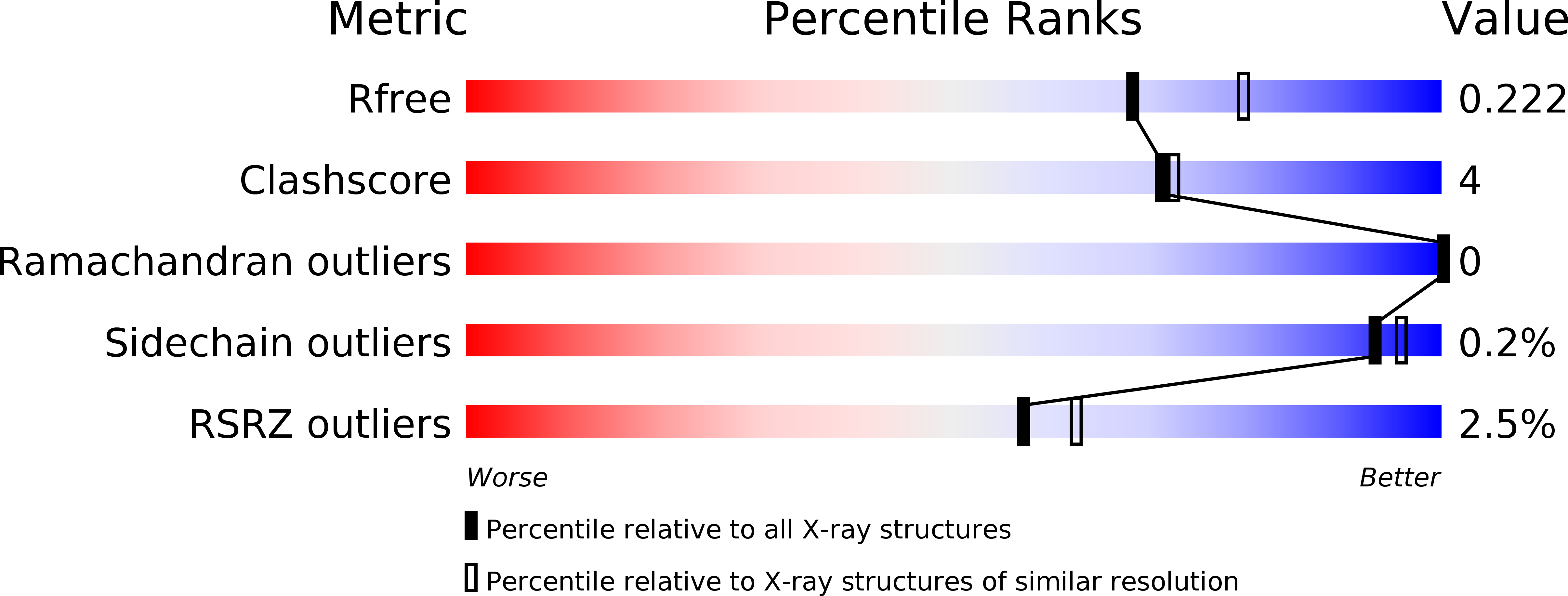
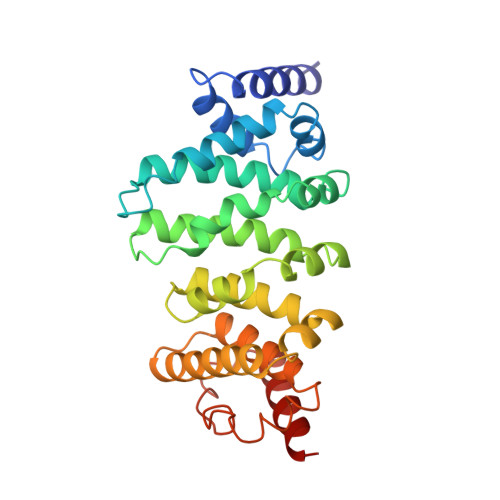
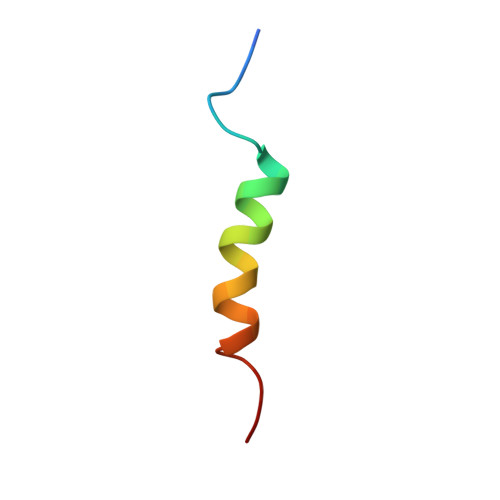
6HOM, 6HON - PubMed Abstract:
The multisubunit CCR4-NOT mRNA deadenylase complex plays important roles in the posttranscriptional regulation of gene expression. The NOT4 E3 ubiquitin ligase is a stable component of the CCR4-NOT complex in yeast but does not copurify with the human or Drosophila melanogaster complex. Here we show that the C-terminal regions of human and D. melanogaster NOT4 contain a conserved sequence motif that directly binds the CAF40 subunit of the CCR4-NOT complex (CAF40-binding motif [CBM]). In addition, nonconserved sequences flanking the CBM also contact other subunits of the complex. Crystal structures of the CBM-CAF40 complex reveal a mutually exclusive binding surface for NOT4 and Roquin or Bag of marbles mRNA regulatory proteins. Furthermore, CAF40 depletion or structure-guided mutagenesis to disrupt the NOT4-CAF40 interaction impairs the ability of NOT4 to elicit decay of tethered reporter mRNAs in cells. Together with additional sequence analyses, our results reveal the molecular basis for the association of metazoan NOT4 with the CCR4-NOT complex and show that it deviates substantially from yeast. They mark the NOT4 ubiquitin ligase as an ancient but nonconstitutive cofactor of the CCR4-NOT deadenylase with potential recruitment and/or effector functions.
Organizational Affiliation:
Department of Biochemistry, Max Planck Institute for Developmental Biology, D-72076 Tübingen, Germany.