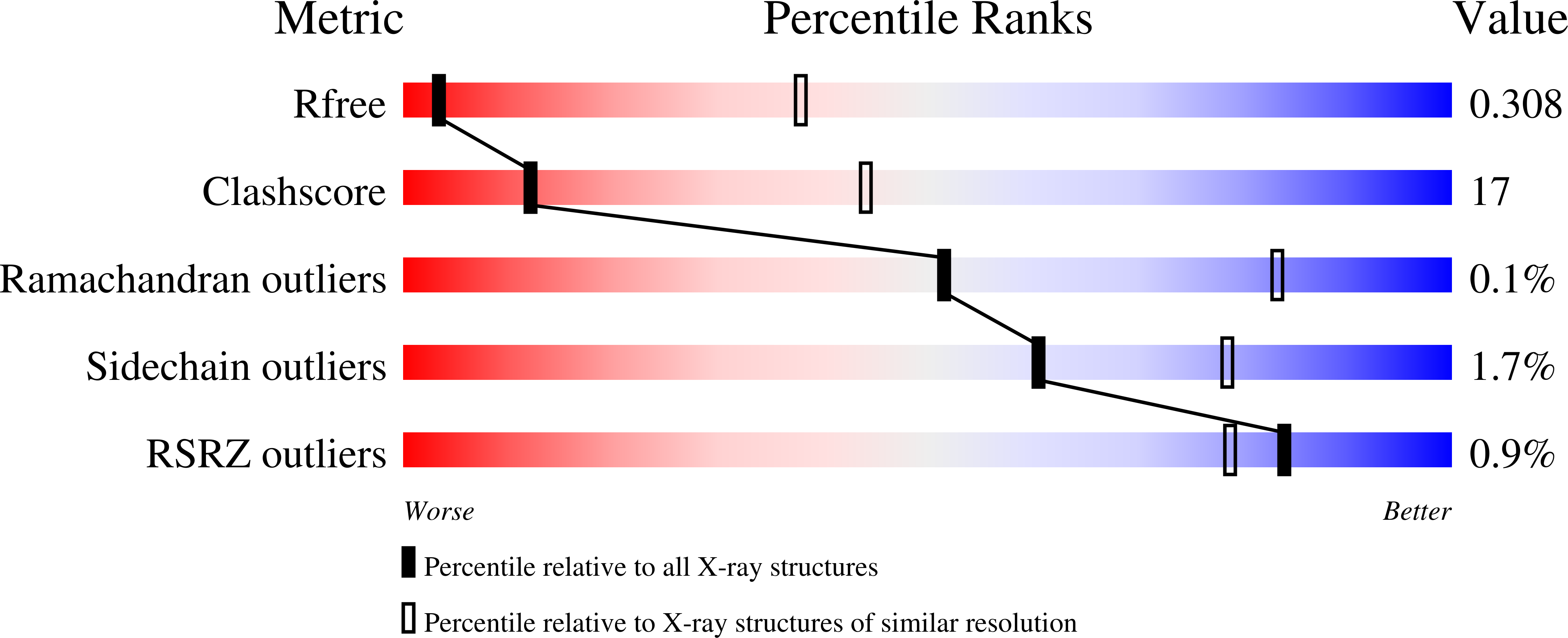
Structural Determinants for Small-Molecule Activation of Skeletal Muscle AMPK alpha 2 beta 2 gamma 1 by the Glucose Importagog SC4.
Ngoei, K.R.W., Langendorf, C.G., Ling, N.X.Y., Hoque, A., Varghese, S., Camerino, M.A., Walker, S.R., Bozikis, Y.E., Dite, T.A., Ovens, A.J., Smiles, W.J., Jacobs, R., Huang, H., Parker, M.W., Scott, J.W., Rider, M.H., Foitzik, R.C., Kemp, B.E., Baell, J.B., Oakhill, J.S.(2018) Cell Chem Biol 25: 728-737.e9
- PubMed: 29657085
- DOI: https://doi.org/10.1016/j.chembiol.2018.03.008
- Primary Citation of Related Structures:
6B1U, 6B2E - PubMed Abstract:
The AMP-activated protein kinase (AMPK) αβγ heterotrimer regulates cellular energy homeostasis with tissue-specific isoform distribution. Small-molecule activation of skeletal muscle α2β2 AMPK complexes may prove a valuable treatment strategy for type 2 diabetes and insulin resistance. Herein, we report the small-molecule SC4 is a potent, direct AMPK activator that preferentially activates α2 complexes and stimulates skeletal muscle glucose uptake. In parallel with the term secretagog, we propose "importagog" to define a substance that induces or augments cellular uptake of another substance. Three-dimensional structures of the glucose importagog SC4 bound to activated α2β2γ1 and α2β1γ1 complexes reveal binding determinants, in particular a key interaction between the SC4 imidazopyridine 4'-nitrogen and β2-Asp111, which provide a design paradigm for β2-AMPK therapeutics. The α2β2γ1/SC4 structure reveals an interaction between a β2 N-terminal α helix and the α2 autoinhibitory domain. Our results provide a structure-function guide to accelerate development of potent, but importantly tissue-specific, β2-AMPK therapeutics.
Organizational Affiliation:
Protein Chemistry & Metabolism, St. Vincent's Institute of Medical Research, University of Melbourne, Fitzroy, VIC 3065, Australia.