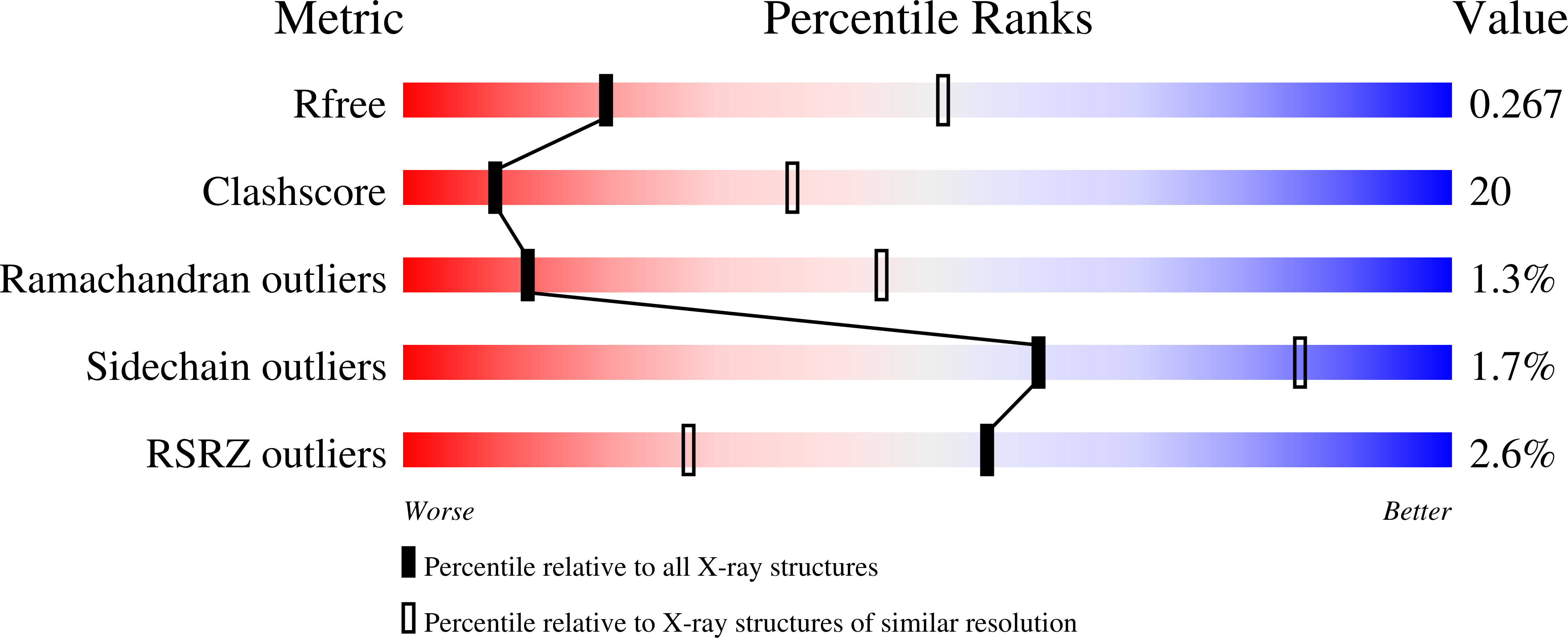
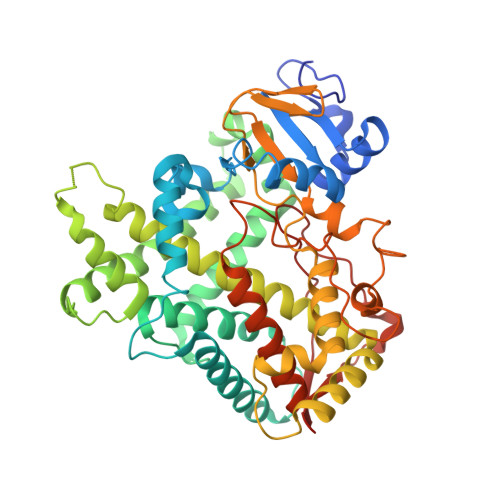
Crystal Structure of CYP2B6 in Complex with an Efavirenz Analog.
Shah, M.B., Zhang, Q., Halpert, J.R.(2018) Int J Mol Sci 19
- PubMed: 29596329
- DOI: https://doi.org/10.3390/ijms19041025
- Primary Citation of Related Structures:
5WBG - PubMed Abstract:
The over two dozen CYP2B structures of human, rabbit, and woodrat enzymes solved in the last decade have significantly enhanced our understanding of the structure-function relationships of drug metabolizing enzymes. More recently, an important role has emerged for halogen-π interactions in the CYP2B6 active site in substrate selectivity, explaining in part the preference for halogenated ligands as substrates. The mechanism by which such ligands interact with CYP2B enzymes involves conserved phenylalanine side chains, in particular F108, F115, or F297, in the active site, which form π bonds with halogens. To illustrate such halogen-π interactions using drugs that are major substrates of CYP2B6, we present here a crystal structure of CYP2B6 in complex with an analog of the widely used anti-HIV drug efavirenz, which contains a methyl group in place of the carbonyl oxygen. The chlorine of the efavirenz analog forms a π bond with the aromatic ring of F108, whereas the putative metabolism site on the distal end of the molecule is oriented towards the heme iron. The crystal structure showcases how CYP2B6 accommodates this important drug analog of considerable size in the active site by movement of various side chains without substantially increasing the active site volume. Furthermore, the CYP2B6-efavirenz analog complex provides a useful platform to investigate computationally as well as biophysically the effect of genetic polymorphisms on binding of the widely studied efavirenz.
Organizational Affiliation:
School of Pharmacy, University of Connecticut, Storrs, CT 06269, USA. manish.shah@acphs.edu.