Comparative structural and mechanistic studies of 4-hydroxy-tetrahydrodipicolinate reductases from Mycobacterium tuberculosis and Vibrio vulnificus.
Pote, S., Kachhap, S., Mank, N.J., Daneshian, L., Klapper, V., Pye, S., Arnette, A.K., Shimizu, L.S., Borowski, T., Chruszcz, M.(2021) Biochim Biophys Acta Gen Subj 1865: 129750-129750
- PubMed: 32980502
- DOI: https://doi.org/10.1016/j.bbagen.2020.129750
- Primary Citation of Related Structures:
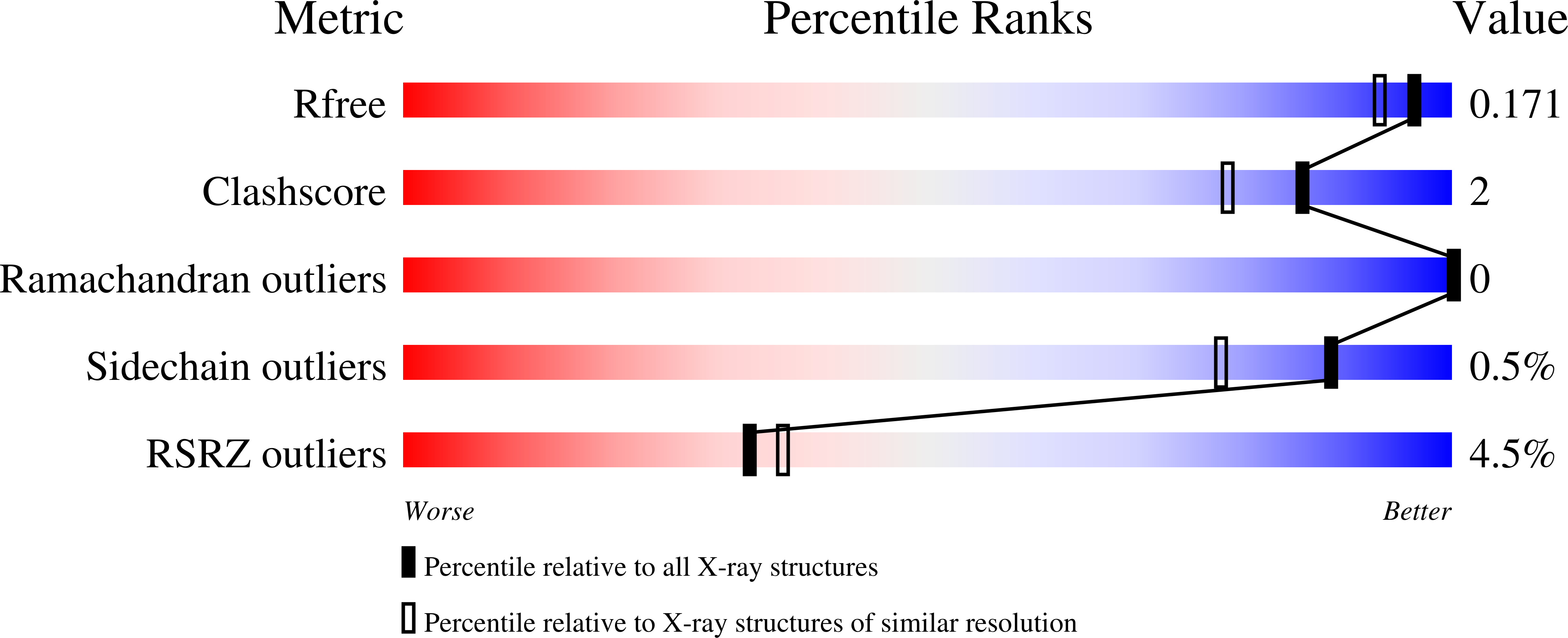
5TEJ, 5TEK, 5TEM, 5TEN, 5TJY, 5TJZ, 5UGV, 5US6 - PubMed Abstract:
The products of the lysine biosynthesis pathway, meso-diaminopimelate and lysine, are essential for bacterial survival. This paper focuses on the structural and mechanistic characterization of 4-hydroxy-tetrahydrodipicolinate reductase (DapB), which is one of the enzymes from the lysine biosynthesis pathway. DapB catalyzes the conversion of (2S, 4S)-4-hydroxy-2,3,4,5-tetrahydrodipicolinate (HTPA) to 2,3,4,5-tetrahydrodipicolinate in an NADH/NADPH dependent reaction. Genes coding for DapBs were identified as essential for many pathogenic bacteria, and therefore DapB is an interesting new target for the development of antibiotics.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of South Carolina, Columbia, SC 29208, USA.