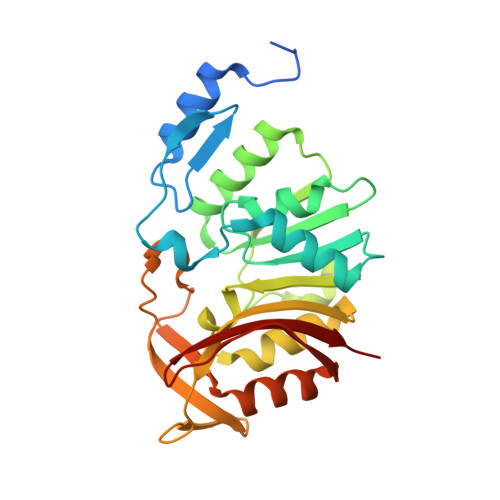
The Structure of the Bifunctional Everninomicin Biosynthetic Enzyme EvdMO1 Suggests Independent Activity of the Fused Methyltransferase-Oxidase Domains.
Starbird, C.A., Perry, N.A., Chen, Q., Berndt, S., Yamakawa, I., Loukachevitch, L.V., Limbrick, E.M., Bachmann, B.O., Iverson, T.M., McCulloch, K.M.(2018) Biochemistry 57: 6827-6837
- PubMed: 30525509
- DOI: https://doi.org/10.1021/acs.biochem.8b00836
- Primary Citation of Related Structures:
5T38, 5T39, 6EC3 - PubMed Abstract:
Members of the orthosomycin family of natural products are decorated polysaccharides with potent antibiotic activity and complex biosynthetic pathways. The defining feature of the orthosomycins is an orthoester linkage between carbohydrate moieties that is necessary for antibiotic activity and is likely formed by a family of conserved oxygenases. Everninomicins are octasaccharide orthosomycins produced by Micromonospora carbonacea that have two orthoester linkages and a methylenedioxy bridge, three features whose formation logically requires oxidative chemistry. Correspondingly, the evd gene cluster encoding everninomicin D encodes two monofunctional nonheme iron, α-ketoglutarate-dependent oxygenases and one bifunctional enzyme with an N-terminal methyltransferase domain and a C-terminal oxygenase domain. To investigate whether the activities of these domains are linked in the bifunctional enzyme EvdMO1, we determined the structure of the N-terminal methyltransferase domain to 1.1 Å and that of the full-length protein to 3.35 Å resolution. Both domains of EvdMO1 adopt the canonical folds of their respective superfamilies and are connected by a short linker. Each domain's active site is oriented such that it faces away from the other domain, and there is no evidence of a channel connecting the two. Our results support EvdMO1 working as a bifunctional enzyme with independent catalytic activities.