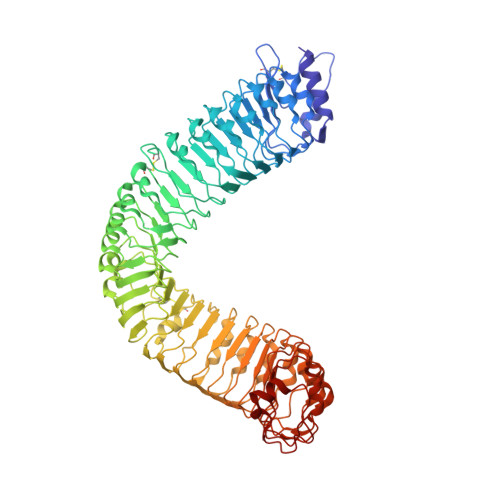
Structural basis for recognition of an endogenous peptide by the plant receptor kinase PEPR1
Tang, J., Han, Z.F., Sun, Y.D., Zhang, H.Q., Gong, X.Q., Chai, J.J.(2015) Cell Res 25: 110-120
- PubMed: 25475059
- DOI: https://doi.org/10.1038/cr.2014.161
- Primary Citation of Related Structures:
5GR8 - PubMed Abstract:
The endogenous peptides AtPep1-8 in Arabidopsis mature from the conserved C-terminal portions of their precursor proteins PROPEP1-8, respectively. The two homologous leucine-rich repeat-receptor kinases (LRR-RKs) PEPR1 and PEPR2 act as receptors of AtPeps. AtPep binding leads to stable association of PEPR1,2 with the shared receptor LRR-RK BAK1, eliciting immune responses similar to those induced by pathogens. Here we report a crystal structure of the extracellular LRR domain of PEPR1 (PEPR1LRR) in complex with AtPep1. The structure reveals that AtPep1 adopts a fully extended conformation and binds to the inner surface of the superhelical PEPR1LRR. Biochemical assays showed that AtPep1 is capable of inducing PEPR1LRR-BAK1LRR heterodimerization. The conserved C-terminal portion of AtPep1 dominates AtPep1 binding to PEPR1LRR, with the last amino acid of AtPep1 Asn23 forming extensive interactions with PEPR1LRR. Deletion of the last residue of AtPep1 significantly compromised AtPep1 interaction with PEPR1LRR. Together, our data reveal a conserved structural mechanism of AtPep1 recognition by PEPR1, providing significant insight into prediction of recognition of other peptides by their cognate LRR-RKs.
Organizational Affiliation:
Tsinghua-Peking Center for Life Sciences, School of Life Sciences, Tsinghua University, Beijing 100084, China.