Radiation Damage and Racemic Protein Crystallography Reveal the Unique Structure of the GASA/Snakin Protein Superfamily.
Yeung, H., Squire, C.J., Yosaatmadja, Y., Panjikar, S., Lopez, G., Molina, A., Baker, E.N., Harris, P.W., Brimble, M.A.(2016) Angew Chem Int Ed Engl 55: 7930-7933
- PubMed: 27145301
- DOI: https://doi.org/10.1002/anie.201602719
- PubMed Abstract:
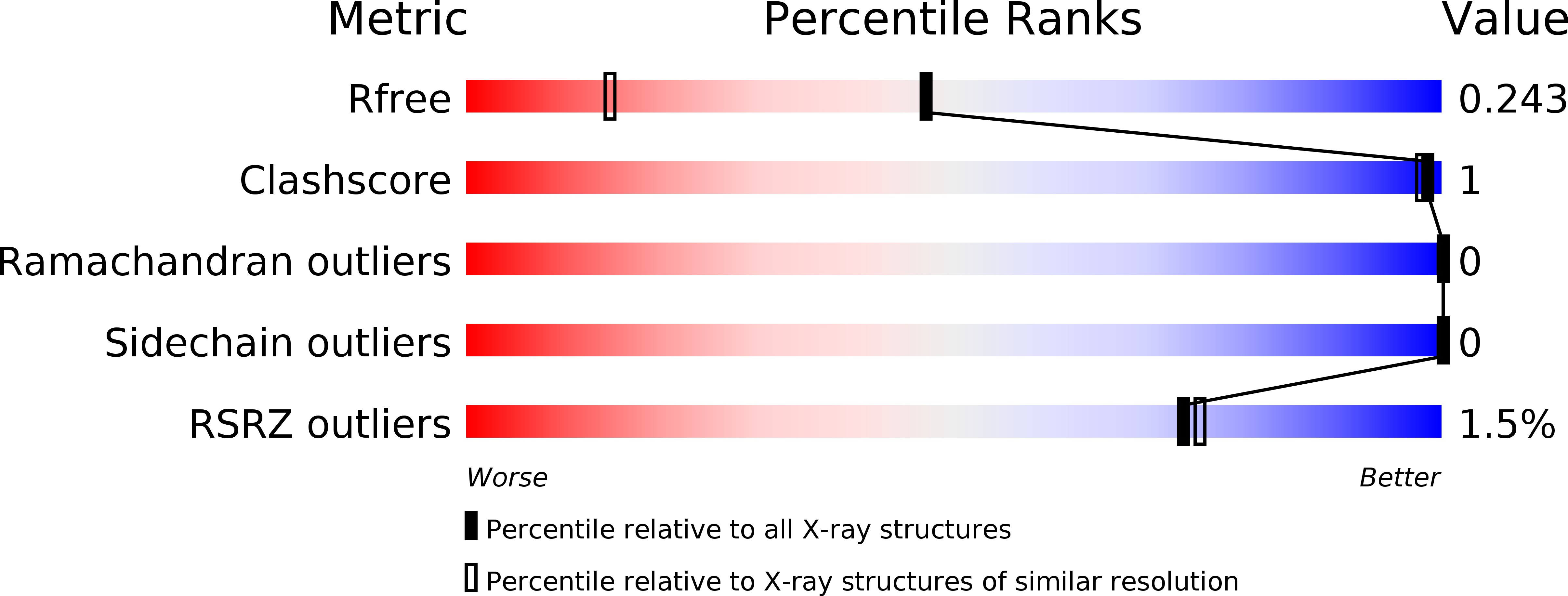
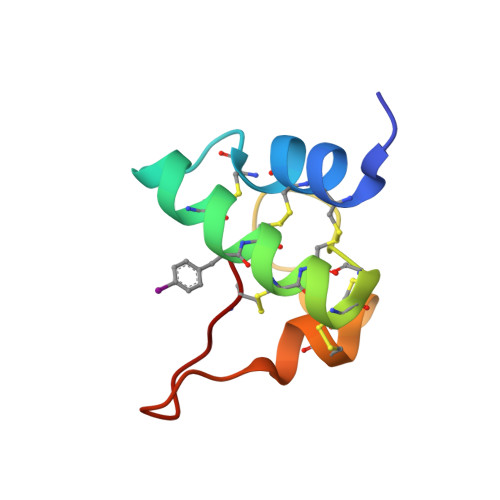
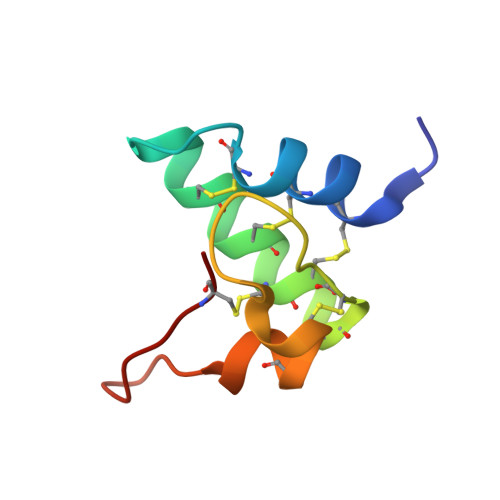
Proteins from the GASA/snakin superfamily are common in plant proteomes and have diverse functions, including hormonal crosstalk, development, and defense. One 63-residue member of this family, snakin-1, an antimicrobial protein from potatoes, has previously been chemically synthesized in a fully active form. Herein the 1.5 Å structure of snakin-1, determined by a novel combination of racemic protein crystallization and radiation-damage-induced phasing (RIP), is reported. Racemic crystals of snakin-1 and quasi-racemic crystals incorporating an unnatural 4-iodophenylalanine residue were prepared from chemically synthesized d- and l-proteins. Breakage of the C-I bonds in the quasi-racemic crystals facilitated structure determination by RIP. The crystal structure reveals a unique protein fold with six disulfide crosslinks, presenting a distinct electrostatic surface that may target the protein to microbial cell surfaces.
Organizational Affiliation:
School of Biological Sciences, The University of Auckland, 3A Symonds St, Auckland Central, 1010, New Zealand.