Mechanistic insight into the nucleus-vacuole junction based on the Vac8p-Nvj1p crystal structure.
Jeong, H., Park, J., Kim, H.I., Lee, M., Ko, Y.J., Lee, S., Jun, Y., Lee, C.(2017) Proc Natl Acad Sci U S A 114: E4539-E4548
- PubMed: 28533415
- DOI: https://doi.org/10.1073/pnas.1701030114
- Primary Citation of Related Structures:
5XJG - PubMed Abstract:
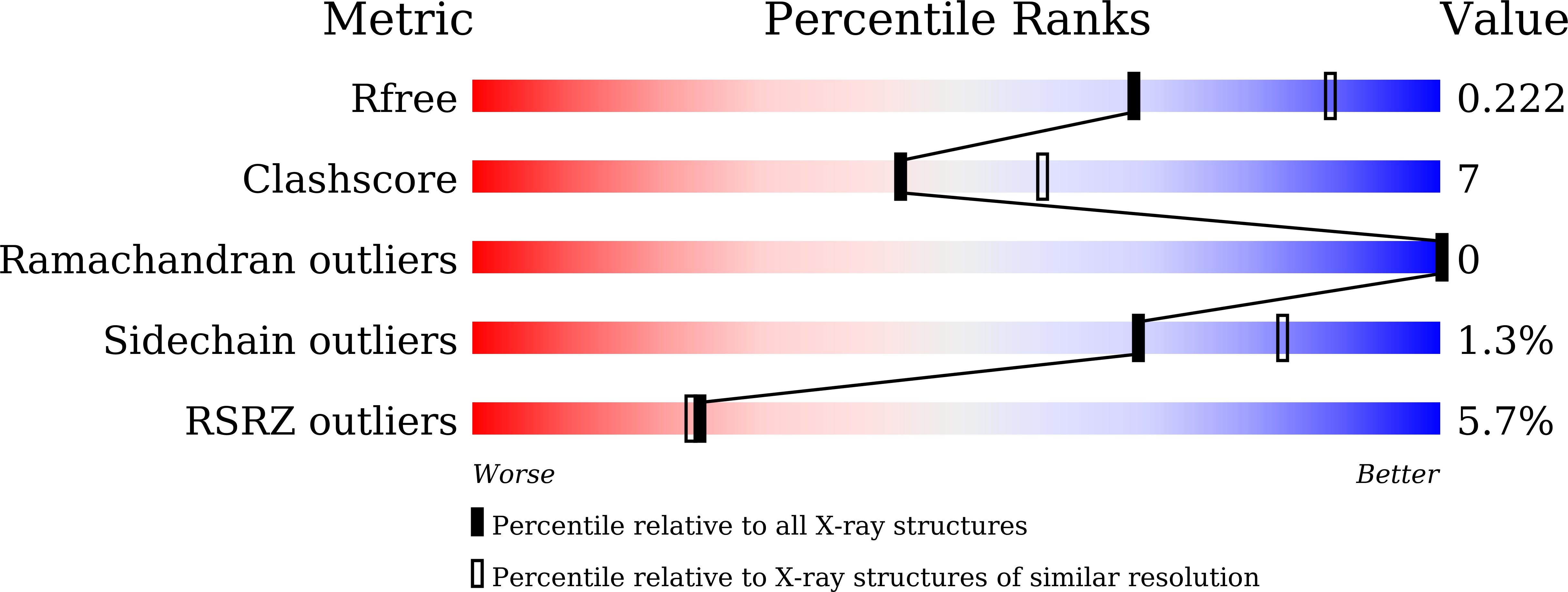
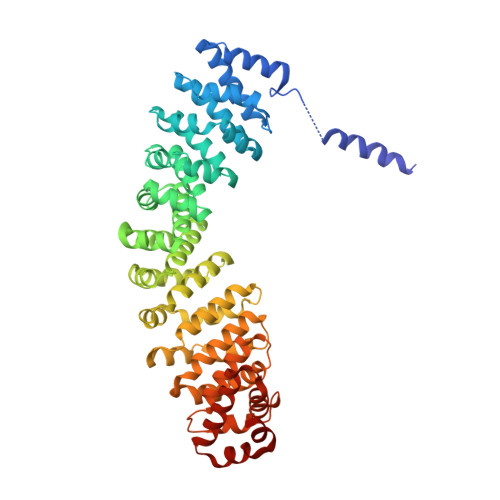
Formation of the nucleus-vacuole junction (NVJ) is mediated by direct interaction between the vacuolar protein Vac8p and the outer nuclear endoplasmic reticulum membrane protein Nvj1p. Herein we report the crystal structure of Vac8p bound to Nvj1p at 2.4-Å resolution. Vac8p comprises a flexibly connected N-terminal H1 helix followed by 12 armadillo repeats (ARMs) that form a right-handed superhelical structure. The extended 80-Å-long loop of Nvj1p specifically binds the highly conserved inner groove formed from ARM1-12 of Vac8p. Disruption of the Nvj1p-Vac8p interaction results in the loss of tight NVJs, which impairs piecemeal microautophagy of the nucleus in Saccharomyces cerevisiae Vac8p cationic triad (Arg276, Arg317, and Arg359) motifs interacting with Nvj1p are also critical to the recognition of Atg13p, a key component of the cytoplasm-to-vacuole targeting (CVT) pathway, indicating competitive binding to Vac8p. Indeed, mutation of the cationic triad abolishes CVT of Ape1p in vivo. Combined with biochemical data, the crystal structure reveals a Vac8p homodimer formed from ARM1, and this self-association, likely regulated by the flexible H1 helix and the C terminus of Nvj1p, is critical for Vac8p cellular functions.
Organizational Affiliation:
Department of Biological Sciences, School of Life Sciences, Ulsan National Institute of Science and Technology, 50 UNIST-gil, Ulsan 44919, Republic of Korea.