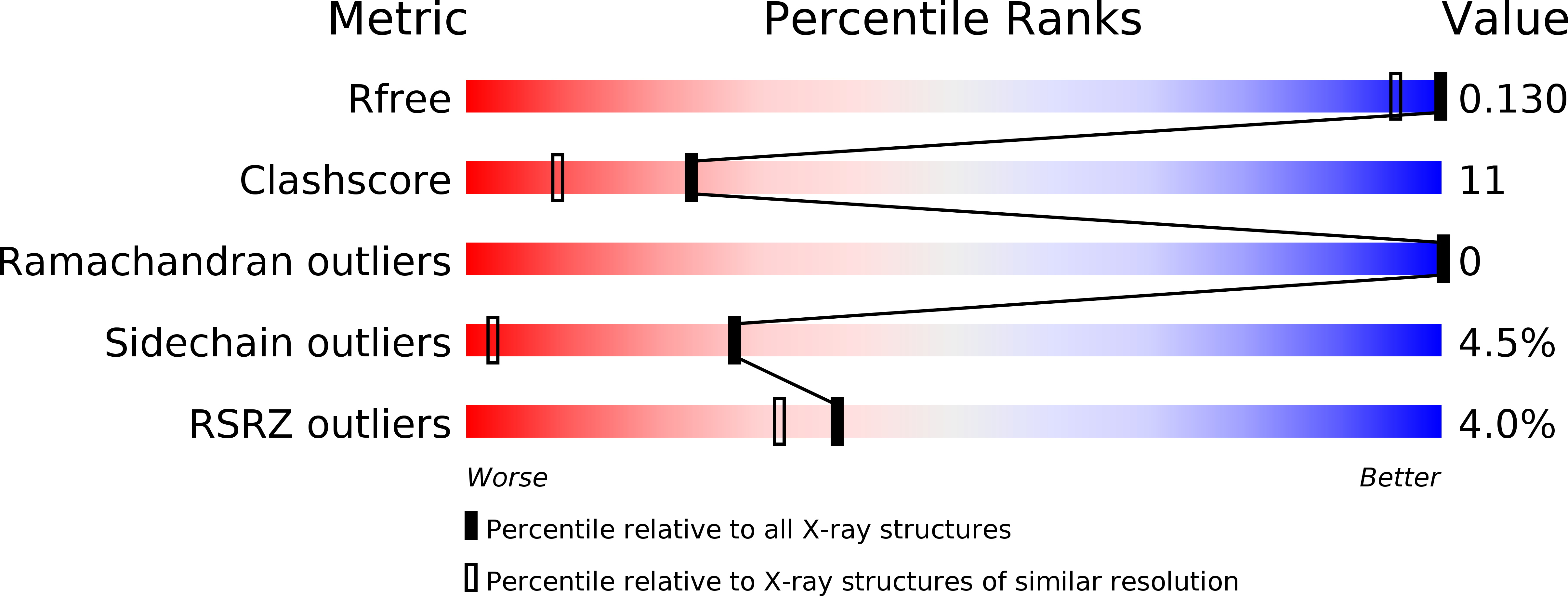
Biosynthesis and genetic encoding of phosphothreonine through parallel selection and deep sequencing.
Zhang, M.S., Brunner, S.F., Huguenin-Dezot, N., Liang, A.D., Schmied, W.H., Rogerson, D.T., Chin, J.W.(2017) Nat Methods 14: 729-736
- PubMed: 28553966
- DOI: https://doi.org/10.1038/nmeth.4302
- Primary Citation of Related Structures:
5NVG - PubMed Abstract:
The phosphorylation of threonine residues in proteins regulates diverse processes in eukaryotic cells, and thousands of threonine phosphorylations have been identified. An understanding of how threonine phosphorylation regulates biological function will be accelerated by general methods to biosynthesize defined phosphoproteins. Here we describe a rapid approach for directly discovering aminoacyl-tRNA synthetase-tRNA pairs that selectively incorporate non-natural amino acids into proteins; our method uses parallel positive selections combined with deep sequencing and statistical analysis and enables the direct, scalable discovery of aminoacyl-tRNA synthetase-tRNA pairs with mutually orthogonal substrate specificity. By combining a method to biosynthesize phosphothreonine in cells with this selection approach, we discover a phosphothreonyl-tRNA synthetase-tRNA CUA pair and create an entirely biosynthetic route to incorporating phosphothreonine in proteins. We biosynthesize several phosphoproteins and demonstrate phosphoprotein structure determination and synthetic protein kinase activation.
Organizational Affiliation:
Medical Research Council Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge, England, UK.