Fe65-PTB2 Dimerization Mimics Fe65-APP Interaction.
Feilen, L.P., Haubrich, K., Strecker, P., Probst, S., Eggert, S., Stier, G., Sinning, I., Konietzko, U., Kins, S., Simon, B., Wild, K.(2017) Front Mol Neurosci 10: 140-140
- PubMed: 28553201
- DOI: https://doi.org/10.3389/fnmol.2017.00140
- Primary Citation of Related Structures:
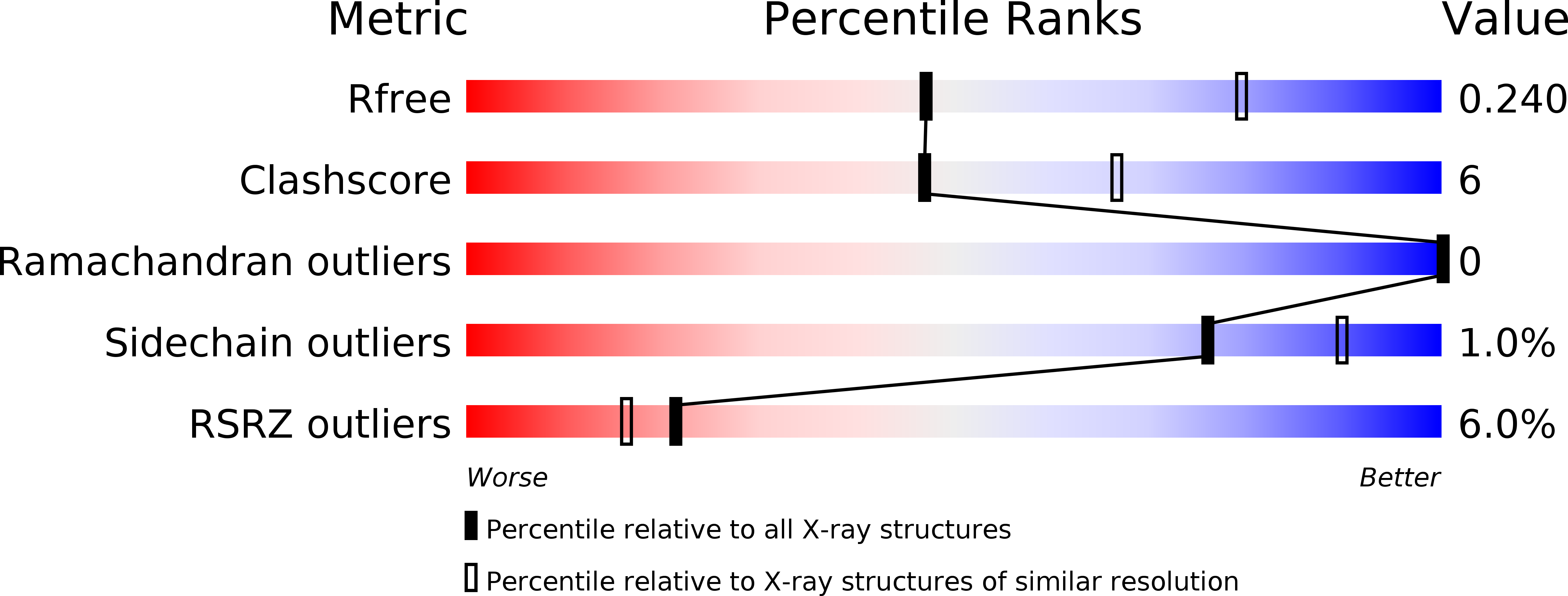
5NQH - PubMed Abstract:
Physiological function and pathology of the Alzheimer's disease causing amyloid precursor protein (APP) are correlated with its cytosolic adaptor Fe65 encompassing a WW and two phosphotyrosine-binding domains (PTBs). The C-terminal Fe65-PTB2 binds a large portion of the APP intracellular domain (AICD) including the GYENPTY internalization sequence fingerprint. AICD binding to Fe65-PTB2 opens an intra-molecular interaction causing a structural change and altering Fe65 activity. Here we show that in the absence of the AICD, Fe65-PTB2 forms a homodimer in solution and determine its crystal structure at 2.6 Å resolution. Dimerization involves the unwinding of a C-terminal α-helix that mimics binding of the AICD internalization sequence, thus shielding the hydrophobic binding pocket. Specific dimer formation is validated by nuclear magnetic resonance (NMR) techniques and cell-based analyses reveal that Fe65-PTB2 together with the WW domain are necessary and sufficient for dimerization. Together, our data demonstrate that Fe65 dimerizes via its APP interaction site, suggesting that besides intra- also intermolecular interactions between Fe65 molecules contribute to homeostatic regulation of APP mediated signaling.
Organizational Affiliation:
Heidelberg University Biochemistry Center (BZH), University of HeidelbergHeidelberg, Germany.