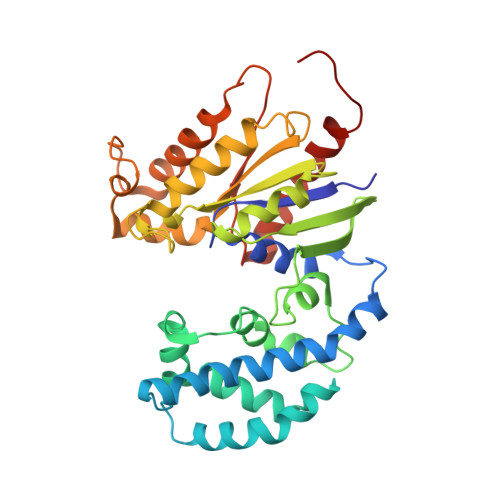
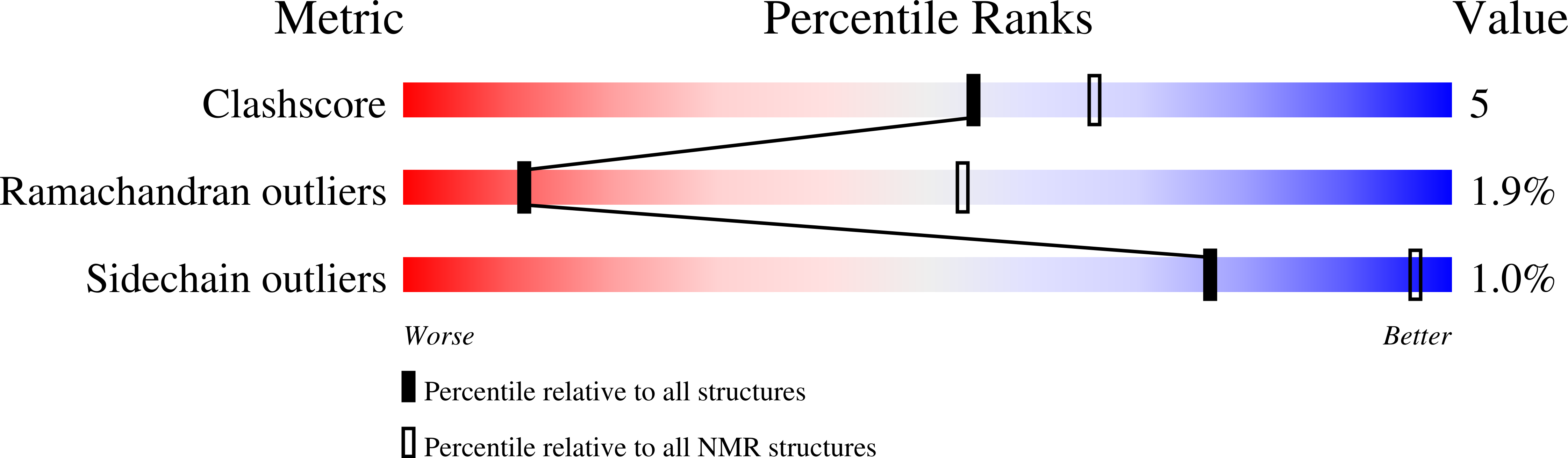Conformational dynamics of a G-protein alpha subunit is tightly regulated by nucleotide binding.
Goricanec, D., Stehle, R., Egloff, P., Grigoriu, S., Pluckthun, A., Wagner, G., Hagn, F.(2016) Proc Natl Acad Sci U S A 113: E3629-E3638
- PubMed: 27298341
- DOI: https://doi.org/10.1073/pnas.1604125113
- Primary Citation of Related Structures:
5JS7, 5JS8 - PubMed Abstract:
Heterotrimeric G proteins play a pivotal role in the signal-transduction pathways initiated by G-protein-coupled receptor (GPCR) activation. Agonist-receptor binding causes GDP-to-GTP exchange and dissociation of the Gα subunit from the heterotrimeric G protein, leading to downstream signaling. Here, we studied the internal mobility of a G-protein α subunit in its apo and nucleotide-bound forms and characterized their dynamical features at multiple time scales using solution NMR, small-angle X-ray scattering, and molecular dynamics simulations. We find that binding of GTP analogs leads to a rigid and closed arrangement of the Gα subdomain, whereas the apo and GDP-bound forms are considerably more open and dynamic. Furthermore, we were able to detect two conformational states of the Gα Ras domain in slow exchange whose populations are regulated by binding to nucleotides and a GPCR. One of these conformational states, the open state, binds to the GPCR; the second conformation, the closed state, shows no interaction with the receptor. Binding to the GPCR stabilizes the open state. This study provides an in-depth analysis of the conformational landscape and the switching function of a G-protein α subunit and the influence of a GPCR in that landscape.
Organizational Affiliation:
Institute for Advanced Study at the Department of Chemistry, Technische Universität München, 85747 Garching, Germany; Center for Integrated Protein Science Munich at the Department of Chemistry, Technische Universität München, 85747 Garching, Germany; Institute of Structural Biology, Helmholtz Zentrum München, 85764 Neuherberg, Germany;