Structural and Functional Study of Apoptosis-linked Gene-2Heme-binding Protein 2 Interactions in HIV-1 Production.
Ma, J., Zhang, X., Feng, Y., Zhang, H., Wang, X., Zheng, Y., Qiao, W., Liu, X.(2016) J Biol Chem 291: 26670-26685
- PubMed: 27784779
- DOI: https://doi.org/10.1074/jbc.M116.752444
- Primary Citation of Related Structures:
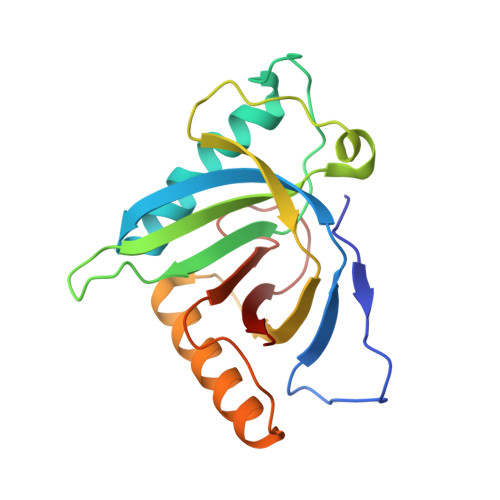
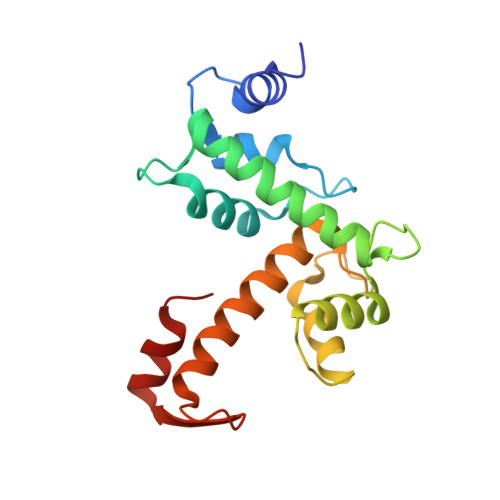
5GQQ - PubMed Abstract:
In the HIV-1 replication cycle, the endosomal sorting complex required for transport (ESCRT) machinery promotes viral budding and release in the late stages. In this process, the ESCRT proteins, ALIX and TSG101, are recruited through interactions with HIV-1 Gag p6. ALG-2, also known as PDCD6, interacts with both ALIX and TSG101 and bridges ESCRT-III and ESCRT-I. In this study, we show that ALG-2 affects HIV-1 production negatively at both the exogenous and endogenous levels. Through a yeast two-hybrid screen, we identified HEBP2 as the binding partner of ALG-2, and we solved the crystal structure of the ALG-2·HEBP2 complex. The function of ALG-2·HEBP2 complex in HIV-1 replication was further explored. ALG-2 inhibits HIV-1 production by affecting Gag expression and distribution, and HEBP2 might aid this process by tethering ALG-2 in the cytoplasm.
Organizational Affiliation:
From the State Key Laboratory of Medicinal Chemical Biology, College of Life Sciences, Nankai University, Tianjin 300071.