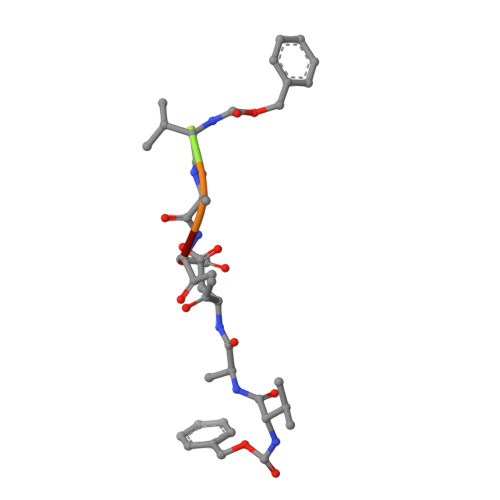
An Unusual Member of the Papain Superfamily: Mapping the Catalytic Cleft of the Marasmius oreades agglutinin (MOA) with a Caspase Inhibitor.
Cordara, G., van Eerde, A., Grahn, E.M., Winter, H.C., Goldstein, I.J., Krengel, U.(2016) PLoS One 11: e0149407-e0149407
- PubMed: 26901797
- DOI: https://doi.org/10.1371/journal.pone.0149407
- Primary Citation of Related Structures:
5D61, 5D62, 5D63 - PubMed Abstract:
Papain-like cysteine proteases (PLCPs) constitute the largest group of thiol-based protein degrading enzymes and are characterized by a highly conserved fold. They are found in bacteria, viruses, plants and animals and involved in a number of physiological and pathological processes, parasitic infections and host defense, making them interesting targets for drug design. The Marasmius oreades agglutinin (MOA) is a blood group B-specific fungal chimerolectin with calcium-dependent proteolytic activity. The proteolytic domain of MOA presents a unique structural arrangement, yet mimicking the main structural elements in known PLCPs. Here we present the X-ray crystal structure of MOA in complex with Z-VAD-fmk, an irreversible caspase inhibitor known to cross-react with PLCPs. The structural data allow modeling of the substrate binding geometry and mapping of the fundamental enzyme-substrate interactions. The new information consolidates MOA as a new, yet strongly atypical member of the papain superfamily. The reported complex is the first published structure of a PLCP in complex with the well characterized caspase inhibitor Z-VAD-fmk.
Organizational Affiliation:
Department of Chemistry, University of Oslo, Oslo, Norway.