Biochemical studies on WbcA, a sugar epimerase from Yersinia enterocolitica.
Salinger, A.J., Brown, H.A., Thoden, J.B., Holden, H.M.(2015) Protein Sci 24: 1633-1639
- PubMed: 26174084
- DOI: https://doi.org/10.1002/pro.2747
- Primary Citation of Related Structures:
5BUV - PubMed Abstract:
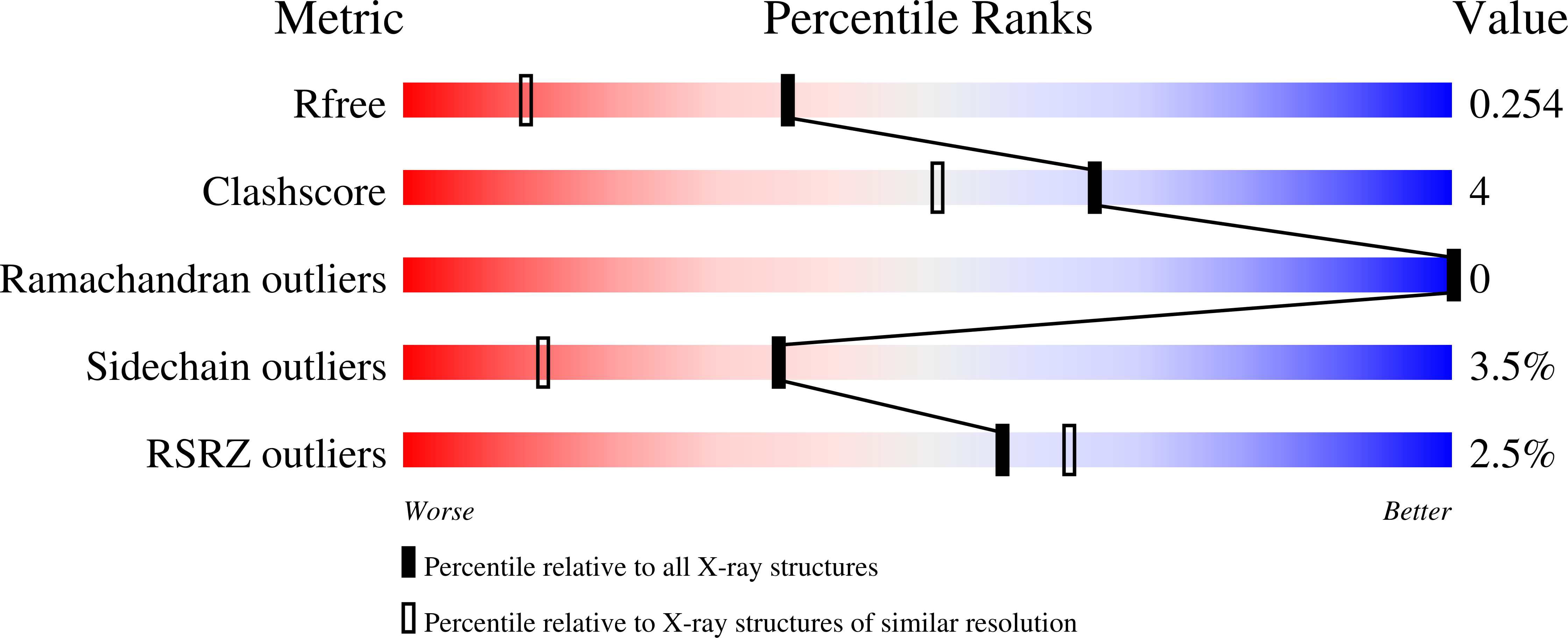
Yersinia enterocolitica is a Gram-negative bacterium that causes yersiniosis, a zoonotic disease affecting the gastrointestinal tract of humans, cattle, and pigs, among others. The lipopolysaccharide of Y. enterocolitica O:8 contains an unusual sugar, 6-deoxy-d-gulose, which requires four enzymes for its biosynthesis. Here, we describe a combined structural and functional investigation of WbcA, which catalyzes the third step in the pathway, namely an epimerization about the C-3' carbon of a CDP-linked sugar. The structure of WbcA was determined to 1.75-Å resolution, and the model was refined to an overall R-factor of 19.5%. The fold of WbcA places it into the well-defined cupin superfamily of sugar epimerases. Typically, these enzymes contain both a conserved histidine and a tyrosine residue that play key roles in catalysis. On the basis of amino acid sequence alignments, it was anticipated that the "conserved" tyrosine had been replaced with a cysteine residue in WbcA (Cys 133), and indeed this was the case. However, what was not anticipated was the fact that another tyrosine residue (Tyr 50) situated on a neighboring β-strand moved into the active site. Site-directed mutant proteins were subsequently constructed and their kinetic properties analyzed to address the roles of Cys 133 and Tyr 50 in WbcA catalysis. This study emphasizes the continuing need to experimentally verify assumptions that are based solely on bioinformatics approaches.
Organizational Affiliation:
Department of Biochemistry, University of Wisconsin, Madison, Wisconsin, 53706.