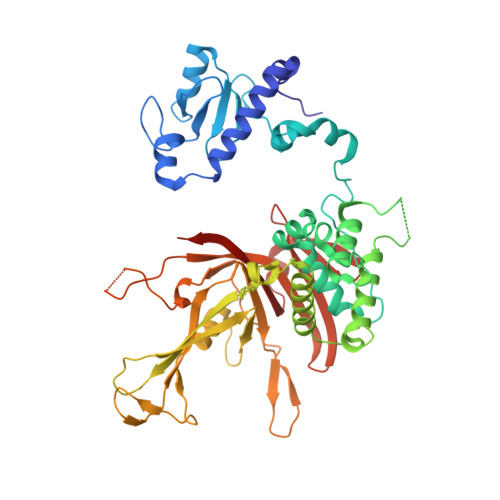
Structural basis for histone H2B deubiquitination by the SAGA DUB module.
Morgan, M.T., Haj-Yahya, M., Ringel, A.E., Bandi, P., Brik, A., Wolberger, C.(2016) Science 351: 725-728
- PubMed: 26912860
- DOI: https://doi.org/10.1126/science.aac5681
- Primary Citation of Related Structures:
4ZUX - PubMed Abstract:
Monoubiquitinated histone H2B plays multiple roles in transcription activation. H2B is deubiquitinated by the Spt-Ada-Gcn5 acetyltransferase (SAGA) coactivator, which contains a four-protein subcomplex known as the deubiquitinating (DUB) module. The crystal structure of the Ubp8/Sgf11/Sus1/Sgf73 DUB module bound to a ubiquitinated nucleosome reveals that the DUB module primarily contacts H2A/H2B, with an arginine cluster on the Sgf11 zinc finger domain docking on the conserved H2A/H2B acidic patch. The Ubp8 catalytic domain mediates additional contacts with H2B, as well as with the conjugated ubiquitin. We find that the DUB module deubiquitinates H2B both in the context of the nucleosome and in H2A/H2B dimers complexed with the histone chaperone, FACT, suggesting that SAGA could target H2B at multiple stages of nucleosome disassembly and reassembly during transcription.
Organizational Affiliation:
Department of Biophysics and Biophysical Chemistry, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.