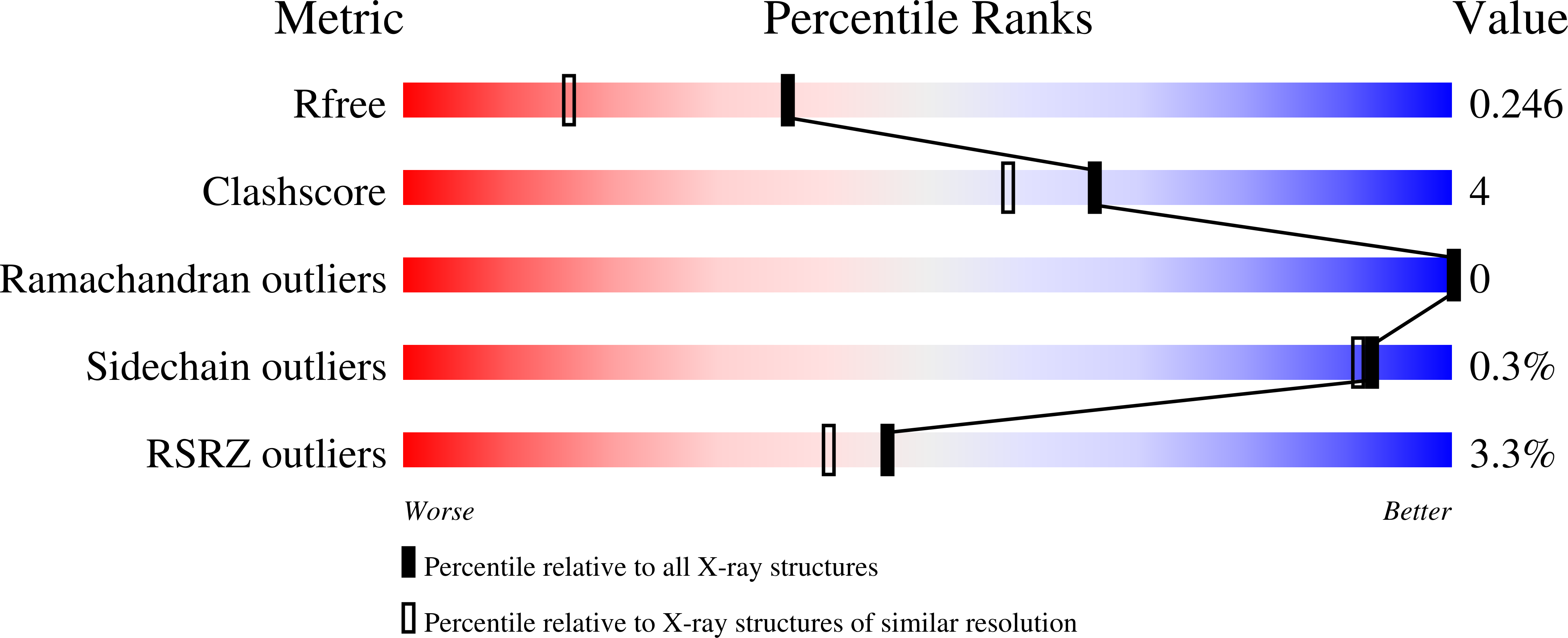
Short Hydrogen Bonds and Proton Delocalization in Green Fluorescent Protein (GFP).
Oltrogge, L.M., Boxer, S.G.(2015) ACS Cent Sci 1: 148-156
- PubMed: 27162964
- DOI: https://doi.org/10.1021/acscentsci.5b00160
- Primary Citation of Related Structures:
4ZF3, 4ZF4, 4ZF5 - PubMed Abstract:
Short hydrogen bonds and specifically low-barrier hydrogen bonds (LBHBs) have been the focus of much attention and controversy for their possible role in enzymatic catalysis. The green fluorescent protein (GFP) mutant S65T, H148D has been found to form a very short hydrogen bond between Asp148 and the chromophore resulting in significant spectral perturbations. Leveraging the unique autocatalytically formed chromophore and its sensitivity to this interaction we explore the consequences of proton affinity matching across this putative LBHB. Through the use of noncanonical amino acids introduced through nonsense suppression or global incorporation, we systematically modify the acidity of the GFP chromophore with halogen substituents. X-ray crystal structures indicated that the length of the interaction with Asp148 is unchanged at ∼2.45 Å while the absorbance spectra demonstrate an unprecedented degree of color tuning with increasing acidity. We utilized spectral isotope effects, isotope fractionation factors, and a simple 1D model of the hydrogen bond coordinate in order to gain insight into the potential energy surface and particularly the role that proton delocalization may play in this putative short hydrogen bond. The data and model suggest that even with the short donor-acceptor distance (∼2.45 Å) and near perfect affinity matching there is not a LBHB, that is, the barrier to proton transfer exceeds the H zero-point energy.
Organizational Affiliation:
Department of Chemistry, Stanford University , Stanford, California 94305-5012, United States.