Molecular pharmacodynamics of emixustat in protection against retinal degeneration.
Zhang, J., Kiser, P.D., Badiee, M., Palczewska, G., Dong, Z., Golczak, M., Tochtrop, G.P., Palczewski, K.(2015) J Clin Invest 125: 2781-2794
- PubMed: 26075817
- DOI: https://doi.org/10.1172/JCI80950
- Primary Citation of Related Structures:
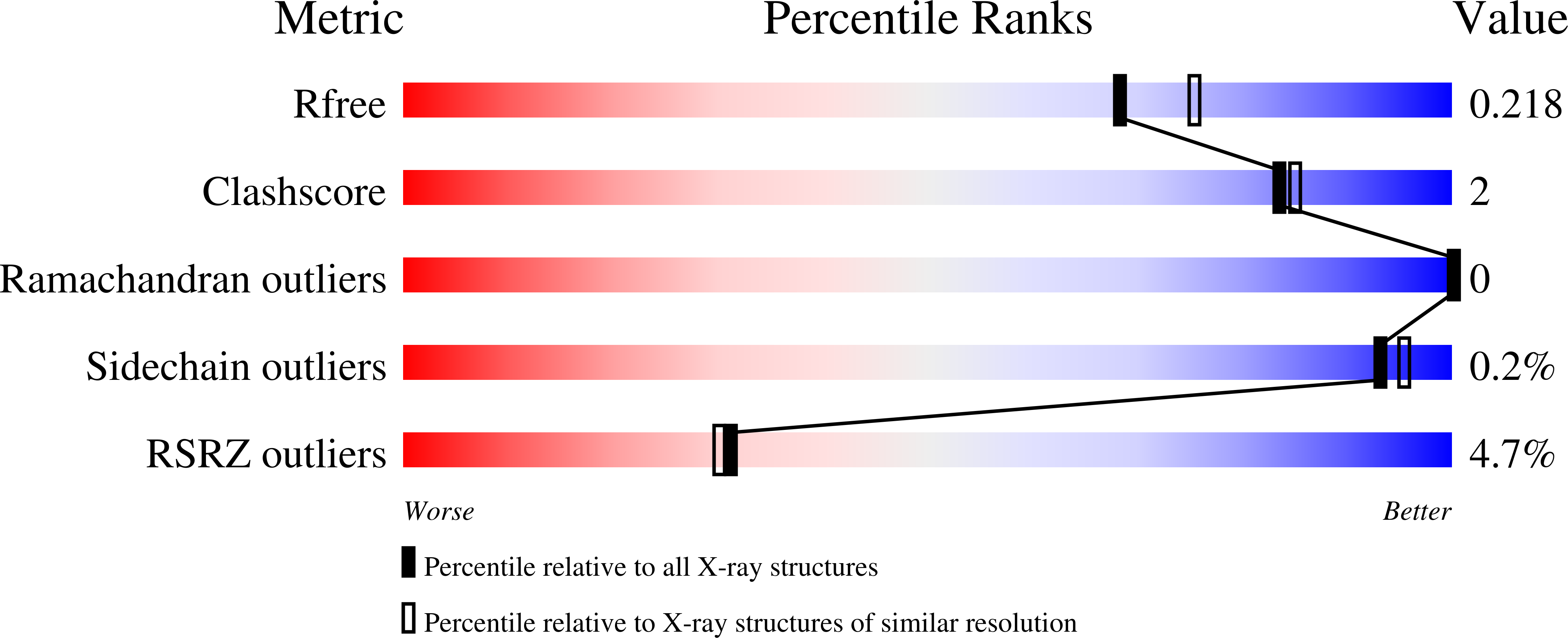
4RYX, 4RYY, 4RYZ, 4ZHK - PubMed Abstract:
Emixustat is a visual cycle modulator that has entered clinical trials as a treatment for age-related macular degeneration (AMD). This molecule has been proposed to inhibit the visual cycle isomerase RPE65, thereby slowing regeneration of 11-cis-retinal and reducing production of retinaldehyde condensation byproducts that may be involved in AMD pathology. Previously, we reported that all-trans-retinal (atRAL) is directly cytotoxic and that certain primary amine compounds that transiently sequester atRAL via Schiff base formation ameliorate retinal degeneration. Here, we have shown that emixustat stereoselectively inhibits RPE65 by direct active site binding. However, we detected the presence of emixustat-atRAL Schiff base conjugates, indicating that emixustat also acts as a retinal scavenger, which may contribute to its therapeutic effects. Using agents that lack either RPE65 inhibitory activity or the capacity to sequester atRAL, we assessed the relative importance of these 2 modes of action in protection against retinal phototoxicity in mice. The atRAL sequestrant QEA-B-001-NH2 conferred protection against phototoxicity without inhibiting RPE65, whereas an emixustat derivative incapable of atRAL sequestration was minimally protective, despite direct inhibition of RPE65. These data indicate that atRAL sequestration is an essential mechanism underlying the protective effects of emixustat and related compounds against retinal phototoxicity. Moreover, atRAL sequestration should be considered in the design of next-generation visual cycle modulators.