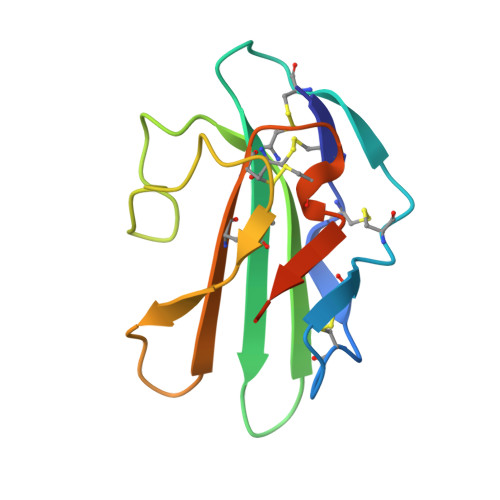
The non-detergent sulfobetaine-201 acts as a pharmacological chaperone to promote folding and crystallization of the type II TGF-beta receptor extracellular domain.
Wangkanont, K., Forest, K.T., Kiessling, L.L.(2015) Protein Expr Purif 115: 19-25
- PubMed: 26073093
- DOI: https://doi.org/10.1016/j.pep.2015.06.001
- Primary Citation of Related Structures:
4P7U - PubMed Abstract:
The roles of the extracellular domain of type II TGF-β receptor (TBRII-ECD) in physiological processes ranging from development to cancer to wound healing render it an attractive target for exploration with chemical tools. For such applications, large amounts of active soluble protein are needed, but the yields of TBRII-ECD we obtained with current folding protocols were variable. To expedite the identification of alternative folding conditions, we developed an on-plate screen. This assay indicated that effective folding additives included the non-detergent sulfobetaine-201 (NDSB-201). Although NDSB-201 can facilitate protein folding, the mode by which it does so is poorly understood. We postulated that specific interactions between NDSB-201 and TBRII-ECD might be responsible. Analysis by X-ray crystallography indicates that the TBRII-ECD possesses a binding pocket for NDSB-201. The pyridinium group of the additive stacks with a phenylalanine side chain in the binding site. The ability of NDSB-201 to occupy a pocket on the protein provides a molecular mechanism for the additive's ability to minimize TBRII-ECD aggregation and stabilize the folded state. NDSB-201 also accelerates TBRII-ECD crystallization, suggesting it may serve as a useful crystallization additive for proteins refolded with it. Our results also suggest there is a site on TBRII-ECD that could be targeted by small-molecule modulators.
Organizational Affiliation:
Department of Chemistry, University of Wisconsin-Madison, 1101 University Avenue, Madison, WI 53706, United States. Electronic address: wangkanont@wisc.edu.