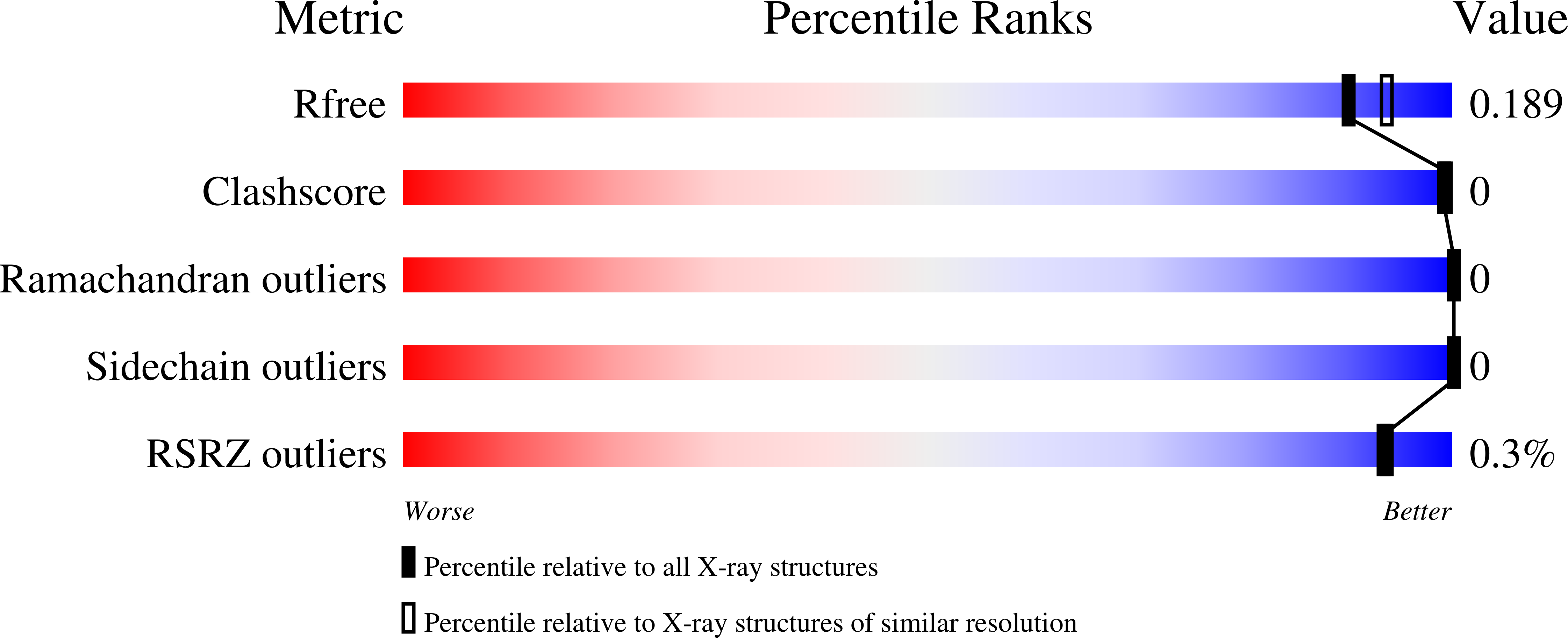
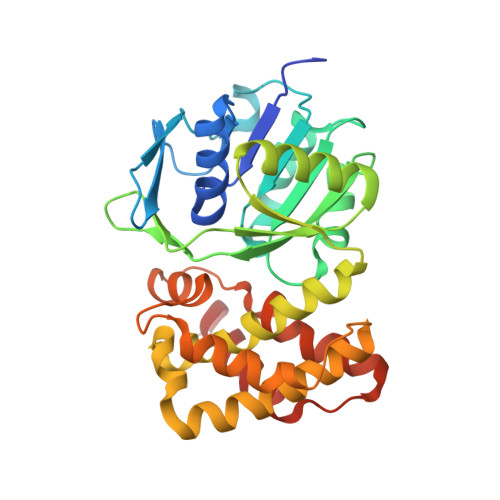
Crystal Structure of putative 2-dehydropantoate 2-reductase PanE from Mycobacterium tuberculosis complexed with NADP and oxamate
Dranow, D.M., Wernimont, A.K., Abendroth, A., Pierce, P.G., Bullen, J., Edwards, T.E., Lorimer, D., Seattle Structural Genomics Center for Infectious DiseaseTo be published.