The co-crystal structure of ubiquitin carboxy-terminal hydrolase L1 (UCHL1) with a tripeptide fluoromethyl ketone (Z-VAE(OMe)-FMK).
Davies, C.W., Chaney, J., Korbel, G., Ringe, D., Petsko, G.A., Ploegh, H., Das, C.(2012) Bioorg Med Chem Lett 22: 3900-3904
- PubMed: 22617491
- DOI: https://doi.org/10.1016/j.bmcl.2012.04.124
- Primary Citation of Related Structures:
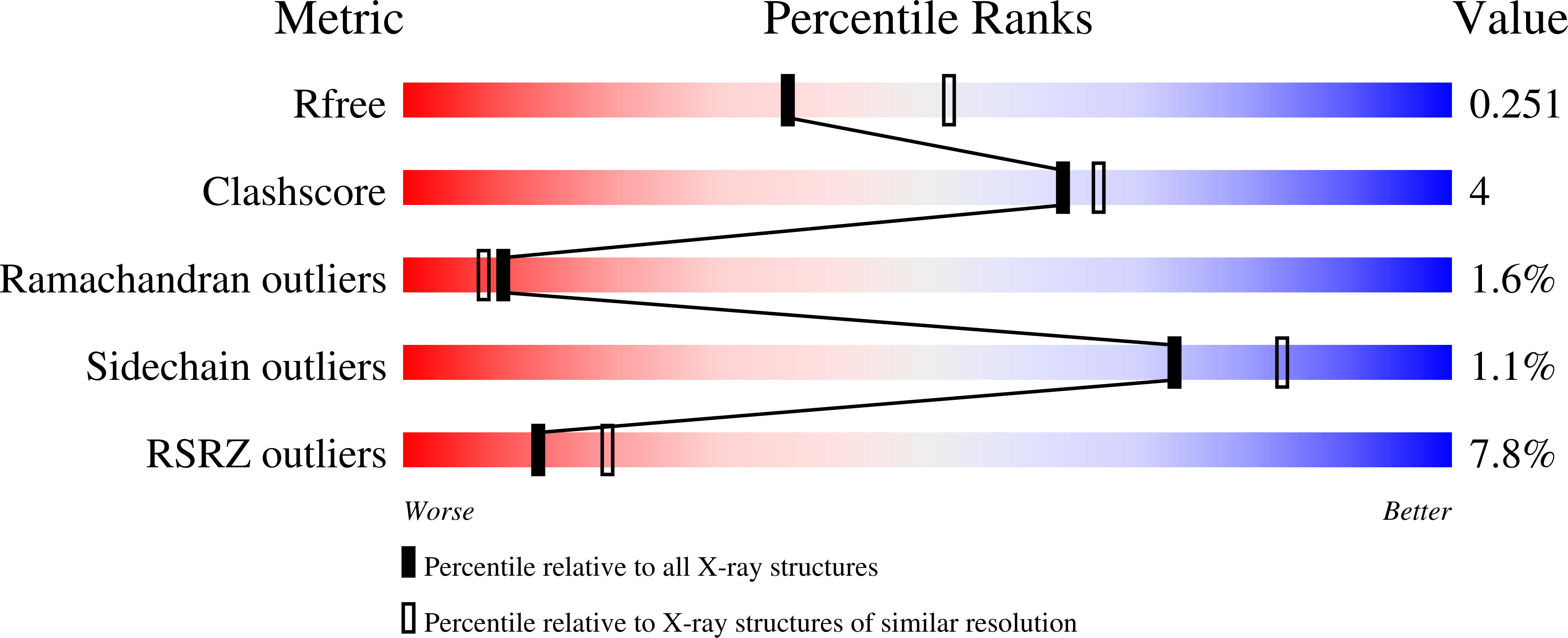
4DM9 - PubMed Abstract:
UCHL1 is a 223 amino acid member of the UCH family of deubiquitinating enzymes (DUBs), found abundantly and exclusively expressed in neurons and the testis in normal tissues. Two naturally occurring variants of UCHL1 are directly involved in Parkinson's disease (PD). Not only has UCHL1 been linked to PD, but it has oncogenic properties, having been found abnormally expressed in lung, pancreatic, and colorectal cancers. Although inhibitors of UCHL1 have been described previously the co-crystal structure of the enzyme bound to any inhibitor has not been reported. Herein, we report the X-ray structure of UCHL1 co-crystallized with a peptide-based fluoromethylketone inhibitor, Z-VAE(OMe)-FMK (VAEFMK) at 2.35 Å resolution. The co-crystal structure reveals that the inhibitor binds in the active-site cleft, irreversibly modifying the active-site cysteine; however, the catalytic histidine is still misaligned as seen in the native structure, suggesting that the inhibitor binds to an inactive form of the enzyme. Our structure also reveals that the inhibitor approaches the active-site cleft from the opposite side of the crossover loop as compared to the direction of approach of ubiquitin's C-terminal tail, thereby occupying the P1' (leaving group) site, a binding site perhaps used by the unknown C-terminal extension of ubiquitin in the actual in vivo substrate(s) of UCHL1. This structure provides a view of molecular contacts at the active-site cleft between the inhibitor and the enzyme as well as furnishing structural information needed to facilitate further design of inhibitors targeted to UCHL1 with high selectivity and potency.
Organizational Affiliation:
Department of Chemistry, Purdue University, West Lafayette, IN 47907, USA.