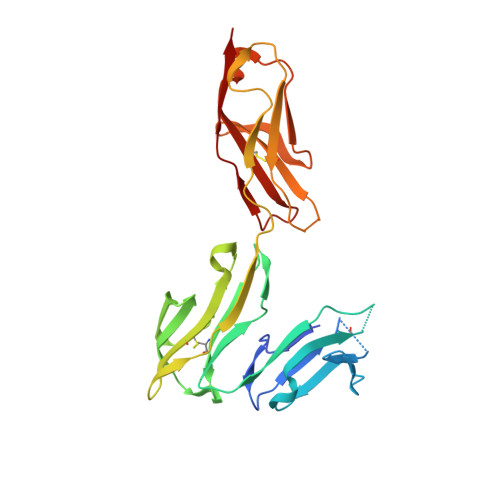
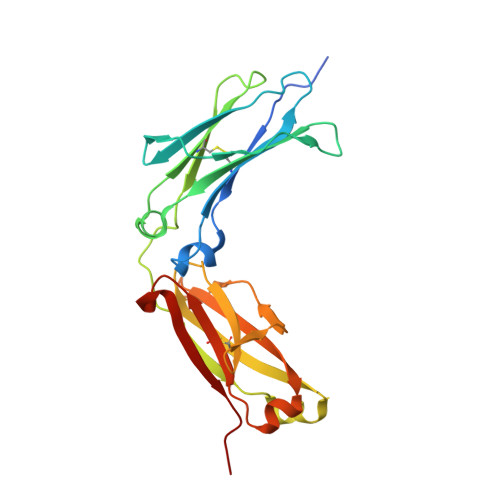
Structural insights into the interaction of human IgG1 with Fc gamma RI: no direct role of glycans in binding.
Oganesyan, V., Mazor, Y., Yang, C., Cook, K.E., Woods, R.M., Ferguson, A., Bowen, M.A., Martin, T., Zhu, J., Wu, H., Dall'Acqua, W.F.(2015) Acta Crystallogr D Biol Crystallogr 71: 2354-2361
- PubMed: 26527150
- DOI: https://doi.org/10.1107/S1399004715018015
- Primary Citation of Related Structures:
4ZNE - PubMed Abstract:
The three-dimensional structure of a human IgG1 Fc fragment bound to wild-type human FcγRI is reported. The structure of the corresponding complex was solved at a resolution of 2.4 Å using molecular replacement; this is the highest resolution achieved for an unmutated FcγRI molecule. This study highlights the critical structural and functional role played by the second extracellular subdomain of FcγRI. It also explains the long-known major energetic contribution of the Fc `LLGG' motif at positions 234-237, and particularly of Leu235, via a `lock-and-key' mechanism. Finally, a previously held belief is corrected and a differing view is offered on the recently proposed direct role of Fc carbohydrates in the corresponding interaction. Structural evidence is provided that such glycan-related effects are strictly indirect.
Organizational Affiliation:
Department of Antibody Discovery and Protein Engineering, MedImmune LLC, 1 MedImmune Way, Gaithersburg, MD 20878, USA.