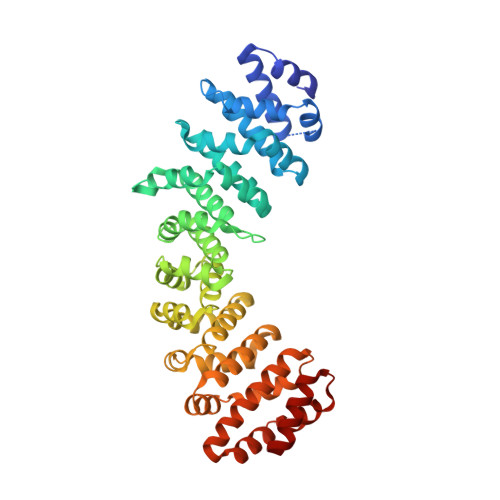
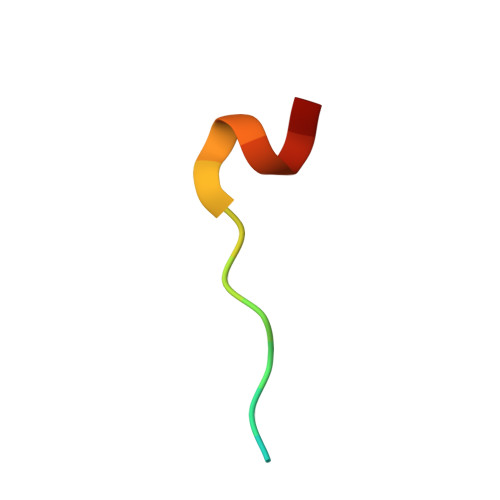
Structure of importin-alpha bound to a non-classical nuclear localization signal of the influenza A virus nucleoprotein
Nakada, R., Hirano, H., Matsuura, Y.(2015) Sci Rep 5: 15055-15055
- PubMed: 26456934
- DOI: https://doi.org/10.1038/srep15055
- Primary Citation of Related Structures:
4ZDU - PubMed Abstract:
A non-classical nuclear localization signal (ncNLS) of influenza A virus nucleoprotein (NP) is critical for nuclear import of viral genomic RNAs that transcribe and replicate in the nucleus of infected cells. Here we report a 2.3 Å resolution crystal structure of mouse importin-α1 in complex with NP ncNLS. The structure reveals that NP ncNLS binds specifically and exclusively to the minor NLS-binding site of importin-α. Structural and functional analyses identify key binding pockets on importin-α as potential targets for antiviral drug development. Unlike many other NLSs, NP ncNLS binds to the NLS-binding domain of importin-α weakly with micromolar affinity. These results suggest that a modest inhibitor with low affinity to importin-α could have anti-influenza activity with minimal cytotoxicity.
Organizational Affiliation:
Division of Biological Science Graduate School of Science Nagoya University Japan.