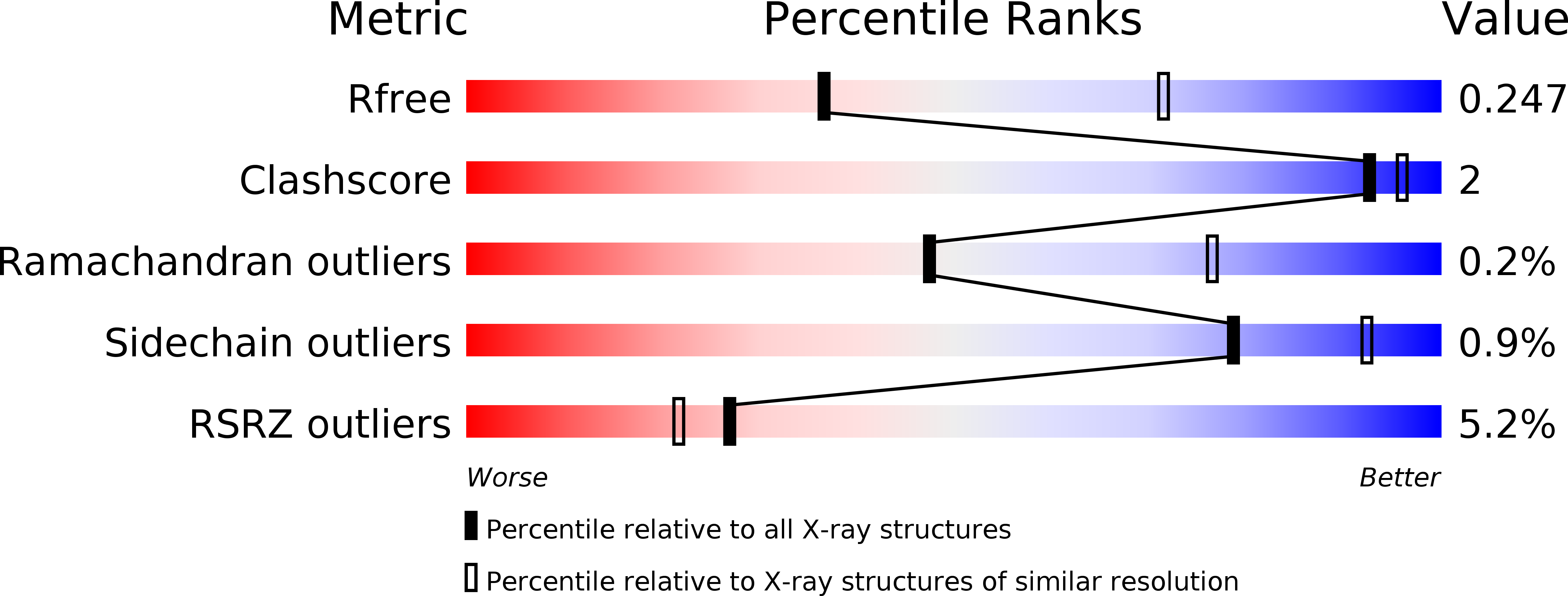
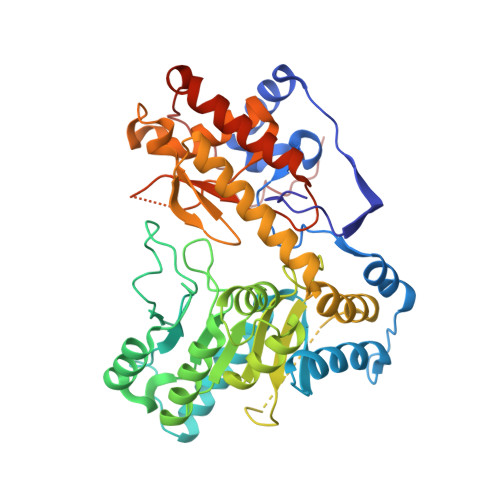
Structure of Escherichia Coli Tryptophanase Purified from an Alkaline-Stressed Bacterial Culture.
Rety, S., Deschamps, P., Leulliot, N.(2015) Acta Crystallogr F Struct Biol Commun 71: 1378
- PubMed: 26527264
- DOI: https://doi.org/10.1107/S2053230X15017549
- Primary Citation of Related Structures:
4UP2 - PubMed Abstract:
Tryptophanase is a bacterial enzyme involved in the degradation of tryptophan to indole, pyruvate and ammonia, which are compounds that are essential for bacterial survival. Tryptophanase is often overexpressed in stressed cultures. Large amounts of endogenous tryptophanase were purified from Escherichia coli BL21 strain overexpressing another recombinant protein. Tryptophanase was crystallized in space group P6522 in the apo form without pyridoxal 5'-phosphate bound in the active site.
Organizational Affiliation:
Laboratoire de Cristallographie et RMN Biologiques, UMR CNRS 8015, Université Paris Descartes, Sorbonne Paris Cité, Faculté de Pharmacie de Paris, Paris, France.