Structural Basis of Human Parechovirus Neutralization by Human Monoclonal Antibodies.
Shakeel, S., Westerhuis, B.M., Ora, A., Koen, G., Bakker, A.Q., Claassen, Y., Wagner, K., Beaumont, T., Wolthers, K.C., Butcher, S.J.(2015) J Virol 89: 9571-9580
- PubMed: 26157123
- DOI: https://doi.org/10.1128/JVI.01429-15
- Primary Citation of Related Structures:
4UDF - PubMed Abstract:
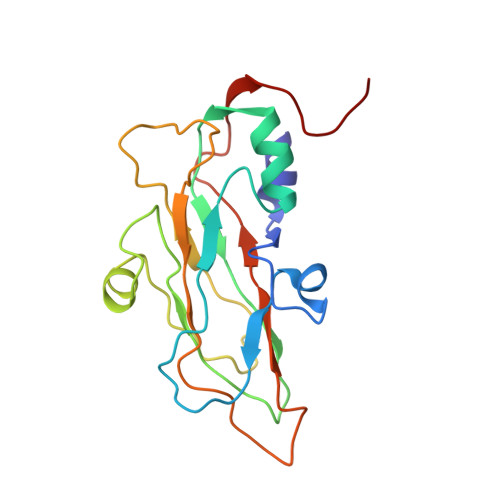
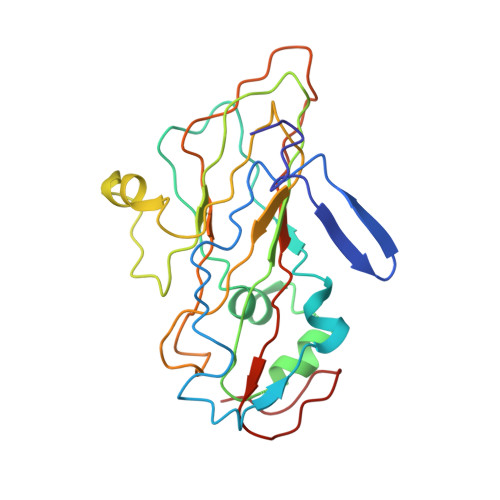
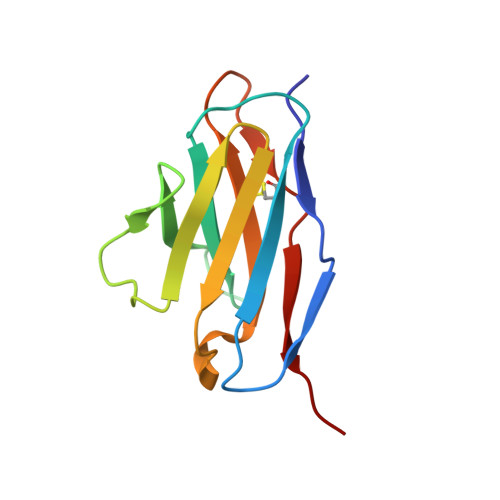
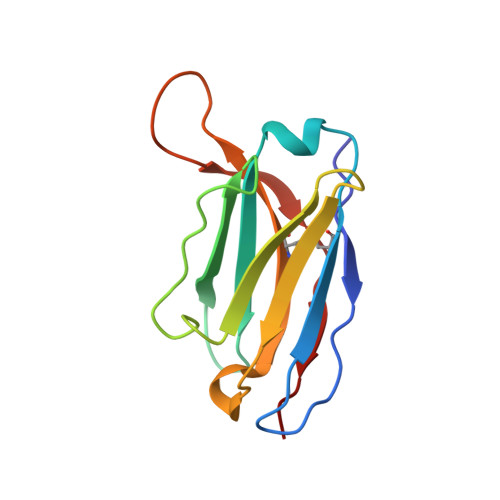
Since it was first recognized in 2004 that human parechoviruses (HPeV) are a significant cause of central nervous system and neonatal sepsis, their clinical importance, primarily in children, has started to emerge. Intravenous immunoglobulin treatment is the only treatment available in such life-threatening cases and has given moderate success. Direct inhibition of parechovirus infection using monoclonal antibodies is a potential treatment. We have developed two neutralizing monoclonal antibodies against HPeV1 and HPeV2, namely, AM18 and AM28, which also cross-neutralize other viruses. Here, we present the mapping of their epitopes using peptide scanning, surface plasmon resonance, fluorescence-based thermal shift assays, electron cryomicroscopy, and image reconstruction. We determined by peptide scanning and surface plasmon resonance that AM18 recognizes a linear epitope motif including the arginine-glycine-aspartic acid on the C terminus of capsid protein VP1. This epitope is normally used by the virus to attach to host cell surface integrins during entry and is found in 3 other viruses that AM18 neutralizes. Therefore, AM18 is likely to cause virus neutralization by aggregation and by blocking integrin binding to the capsid. Further, we show by electron cryomicroscopy, three-dimensional reconstruction, and pseudoatomic model fitting that ordered RNA interacts with HPeV1 VP1 and VP3. AM28 recognizes quaternary epitopes on the capsid composed of VP0 and VP3 loops from neighboring pentamers, thereby increasing the RNA accessibility temperature for the virus-AM28 complex compared to the virus alone. Thus, inhibition of RNA uncoating probably contributes to neutralization by AM28.
Organizational Affiliation:
Institute of Biotechnology, University of Helsinki, Helsinki, Finland.