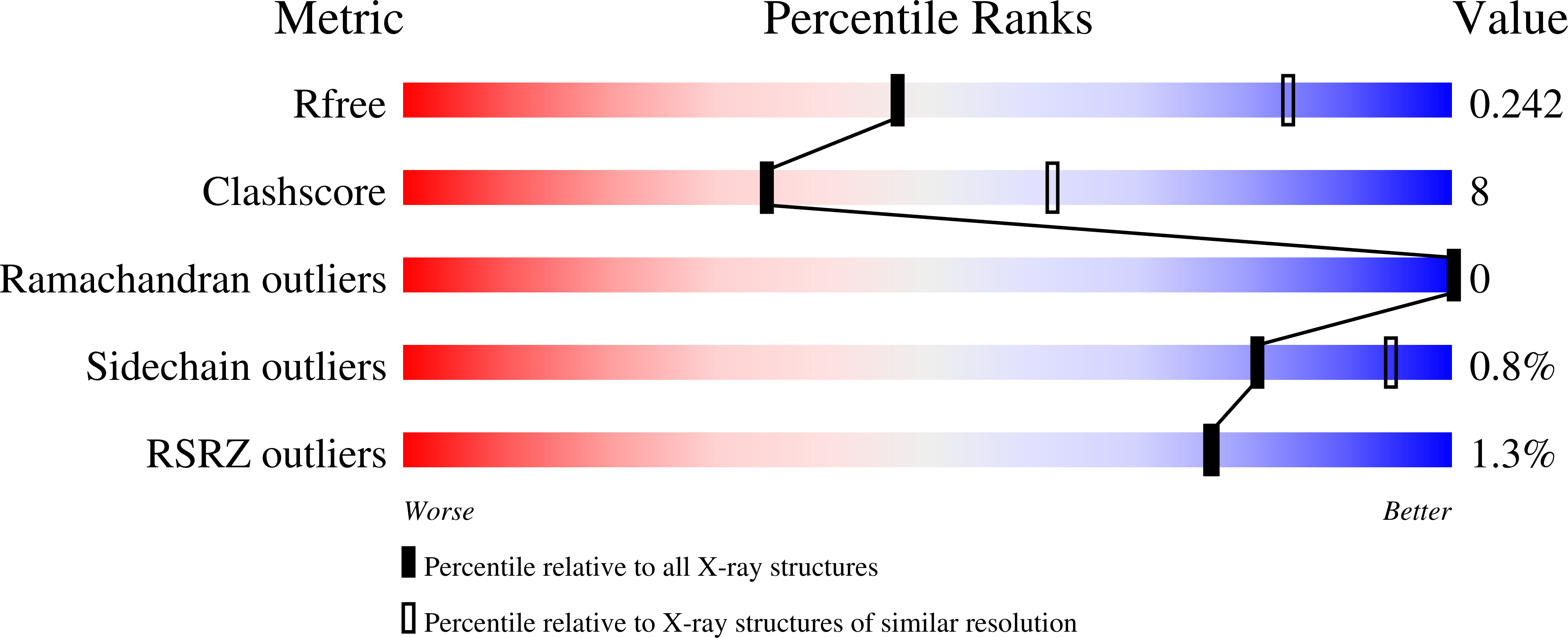
Structure of an SspH1-PKN1 Complex Reveals the Basis for Host Substrate Recognition and Mechanism of Activation for a Bacterial E3 Ubiquitin Ligase.
Keszei, A.F., Tang, X., McCormick, C., Zeqiraj, E., Rohde, J.R., Tyers, M., Sicheri, F.(2014) Mol Cell Biol 34: 362-373
- PubMed: 24248594
- DOI: https://doi.org/10.1128/MCB.01360-13
- Primary Citation of Related Structures:
4NKG, 4NKH - PubMed Abstract:
IpaH proteins are bacterium-specific E3 enzymes that function as type three secretion system (T3SS) effectors in Salmonella, Shigella, and other Gram-negative bacteria. IpaH enzymes recruit host substrates for ubiquitination via a leucine-rich repeat (LRR) domain, which can inhibit the catalytic domain in the absence of substrate. The basis for substrate recognition and the alleviation of autoinhibition upon substrate binding is unknown. Here, we report the X-ray structure of Salmonella SspH1 in complex with human PKN1. The LRR domain of SspH1 interacts specifically with the HR1b coiled-coil subdomain of PKN1 in a manner that sterically displaces the catalytic domain from the LRR domain, thereby activating catalytic function. SspH1 catalyzes the ubiquitination and proteasome-dependent degradation of PKN1 in cells, which attenuates androgen receptor responsiveness but not NF-κB activity. These regulatory features are conserved in other IpaH-substrate interactions. Our results explain the mechanism whereby substrate recognition and enzyme autoregulation are coupled in this class of bacterial ubiquitin ligases.
Organizational Affiliation:
Lunenfeld-Tanenbaum Research Institute, Mount Sinai Hospital, Toronto, Ontario, Canada.