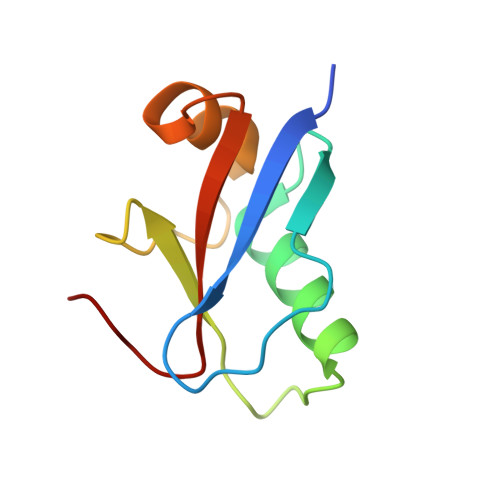
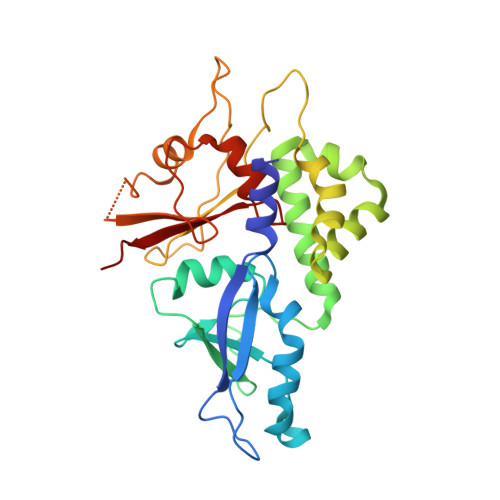
Structural basis of ATG3 recognition by the autophagic ubiquitin-like protein ATG12.
Metlagel, Z., Otomo, C., Takaesu, G., Otomo, T.(2013) Proc Natl Acad Sci U S A 110: 18844-18849
- PubMed: 24191030
- DOI: https://doi.org/10.1073/pnas.1314755110
- Primary Citation of Related Structures:
4NAW - PubMed Abstract:
The autophagic ubiquitin-like protein (ublp) autophagy-related (ATG)12 is a component of the ATG12∼ATG5-ATG16L1 E3 complex that promotes lipid conjugation of members of the LC3 ublp family. A role of ATG12 in the E3 complex is to recruit the E2 enzyme ATG3. Here we report the identification of the ATG12 binding sequence in the flexible region of human ATG3 and the crystal structure of the minimal E3 complexed with the identified binding fragment of ATG3. The structure shows that 13 residues of the ATG3 fragment form a short β-strand followed by an α-helix on a surface area that is exclusive to ATG12. Mutational analyses of ATG3 confirm that four residues whose side chains make contacts with ATG12 are important for E3 interaction as well as LC3 lipidation. Conservation of these four critical residues is high in metazoan organisms and plants but lower in fungi. A structural comparison reveals that the ATG3 binding surface on ATG12 contains a hydrophobic pocket corresponding to the binding pocket of LC3 that accommodates the leucine of the LC3-interacting region motif. These findings establish the mechanism of ATG3 recruitment by ATG12 in higher eukaryotes and place ATG12 among the members of signaling ublps that bind liner sequences.
Organizational Affiliation:
Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037.