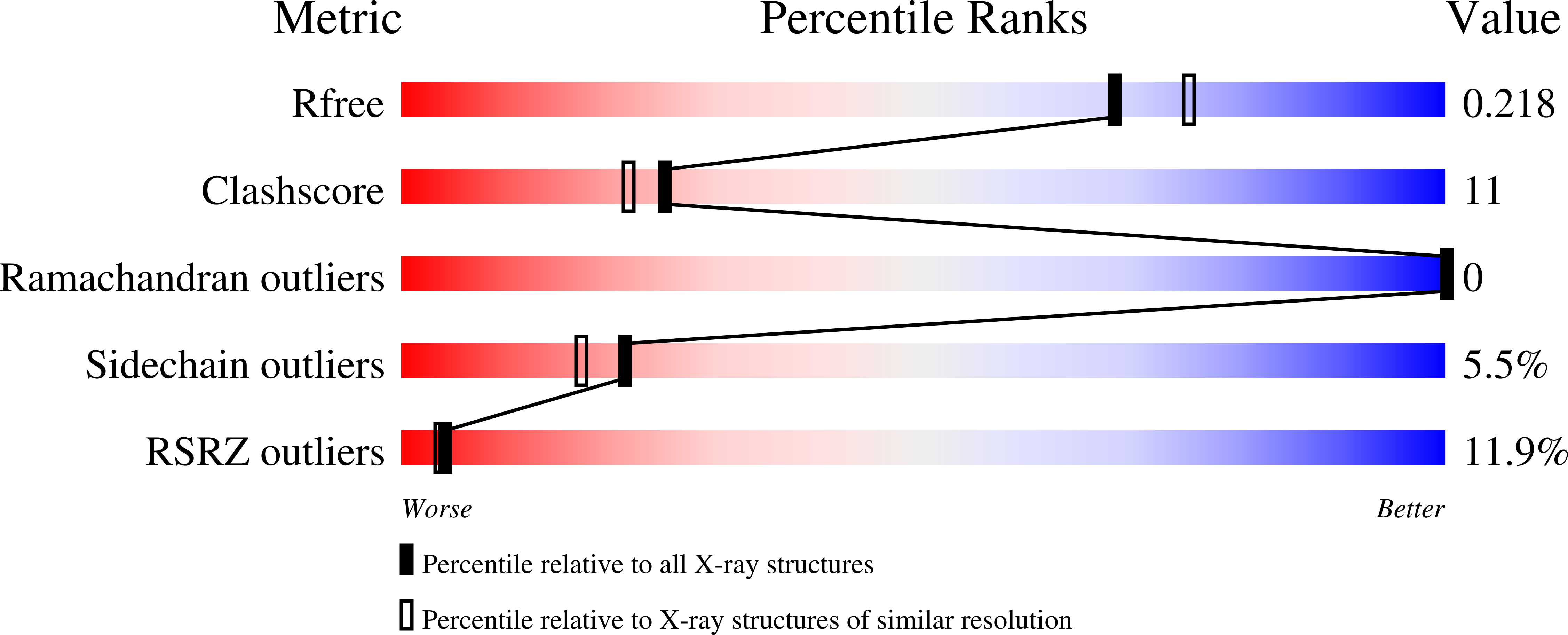
Structural and biochemical basis for the inhibition of cell death by APIP, a methionine salvage enzyme.
Kang, W., Hong, S.H., Lee, H.M., Kim, N.Y., Lim, Y.C., Le, L.T.M., Lim, B., Kim, H.C., Kim, T.Y., Ashida, H., Yokota, A., Hah, S.S., Chun, K.H., Jung, Y.K., Yang, J.K.(2014) Proc Natl Acad Sci U S A 111: 581-584
- PubMed: 24367089
- DOI: https://doi.org/10.1073/pnas.1308768111
- Primary Citation of Related Structures:
4M6R - PubMed Abstract:
APIP, Apaf-1 interacting protein, has been known to inhibit two main types of programmed cell death, apoptosis and pyroptosis, and was recently found to be associated with cancers and inflammatory diseases. Distinct from its inhibitory role in cell death, APIP was also shown to act as a 5-methylthioribulose-1-phosphate dehydratase, or MtnB, in the methionine salvage pathway. Here we report the structural and enzymatic characterization of human APIP as an MtnB enzyme with a Km of 9.32 μM and a Vmax of 1.39 μmol min(-1) mg(-1). The crystal structure was determined at 2.0-Å resolution, revealing an overall fold similar to members of the zinc-dependent class II aldolase family. APIP/MtnB exists as a tetramer in solution and exhibits an assembly with C4 symmetry in the crystal lattice. The pocket-shaped active site is located at the end of a long cleft between two adjacent subunits. We propose an enzymatic reaction mechanism involving Glu139* as a catalytic acid/base, as supported by enzymatic assay, substrate-docking study, and sequence conservation analysis. We explored the relationship between two distinct functions of APIP/MtnB, cell death inhibition, and methionine salvage, by measuring the ability of enzymatic mutants to inhibit cell death, and determined that APIP/MtnB functions as a cell death inhibitor independently of its MtnB enzyme activity for apoptosis induced by either hypoxia or etoposide, but dependently for caspase-1-induced pyroptosis. Our results establish the structural and biochemical groundwork for future mechanistic studies of the role of APIP/MtnB in modulating cell death and inflammation and in the development of related diseases.
Organizational Affiliation:
Department of Chemistry, College of Natural Sciences, Soongsil University, Seoul 156-743, Korea.