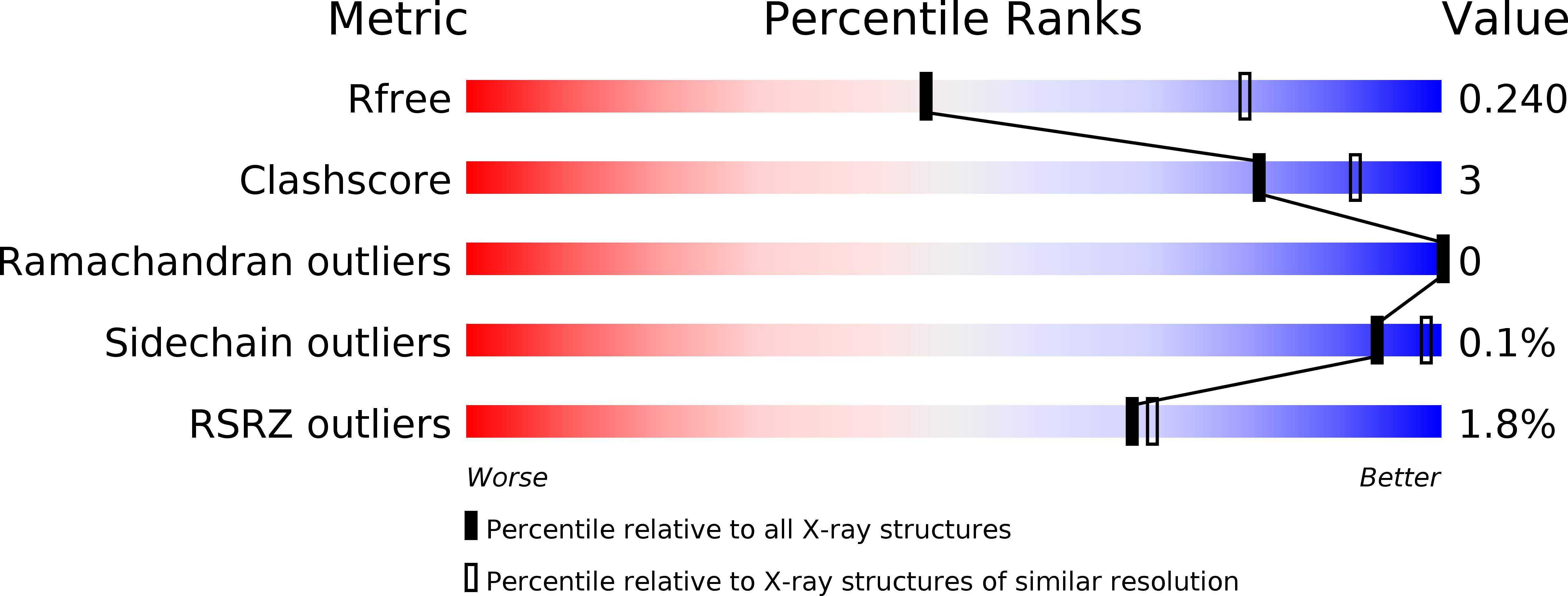
Structure of the human ATG12~ATG5 conjugate required for LC3 lipidation in autophagy.
Otomo, C., Metlagel, Z., Takaesu, G., Otomo, T.(2013) Nat Struct Mol Biol 20: 59-66
- PubMed: 23202584
- DOI: https://doi.org/10.1038/nsmb.2431
- Primary Citation of Related Structures:
4GDK, 4GDL - PubMed Abstract:
The autophagy factor ATG12~ATG5 conjugate exhibits E3 ligase-like activity which facilitates the lipidation of members of the LC3 family. The crystal structure of the human ATG12~ATG5 conjugate bound to the N-terminal region of ATG16L1, the factor that recruits the conjugate to autophagosomal membranes, reveals an integrated architecture in which ATG12 docks onto ATG5 through conserved residues. ATG12 and ATG5 are oriented such that other conserved residues on each molecule, including the conjugation junction, form a continuous surface patch. Mutagenesis data support the importance of both the interface between ATG12 and ATG5 and the continuous patch for E3 activity. The ATG12~ATG5 conjugate interacts with the E2 enzyme ATG3 with high affinity through another surface location that is exclusive to ATG12, suggesting a different role of the continuous patch in E3 activity. These findings provide a foundation for understanding the mechanism of LC3 lipidation.
Organizational Affiliation:
Department of Molecular Biology, The Scripps Research Institute, La Jolla, California, USA.