Structural basis for membrane binding specificity of the Bin/Amphiphysin/Rvs (BAR) domain of Arfaptin-2 determined by Arl1 GTPase
Nakamura, K., Man, Z., Xie, Y., Hanai, A., Makyio, H., Kawasaki, M., Kato, R., Shin, H.-W., Nakayama, K., Wakatsuki, S.(2012) J Biol Chem 287: 25478-25489
- PubMed: 22679020
- DOI: https://doi.org/10.1074/jbc.M112.365783
- Primary Citation of Related Structures:
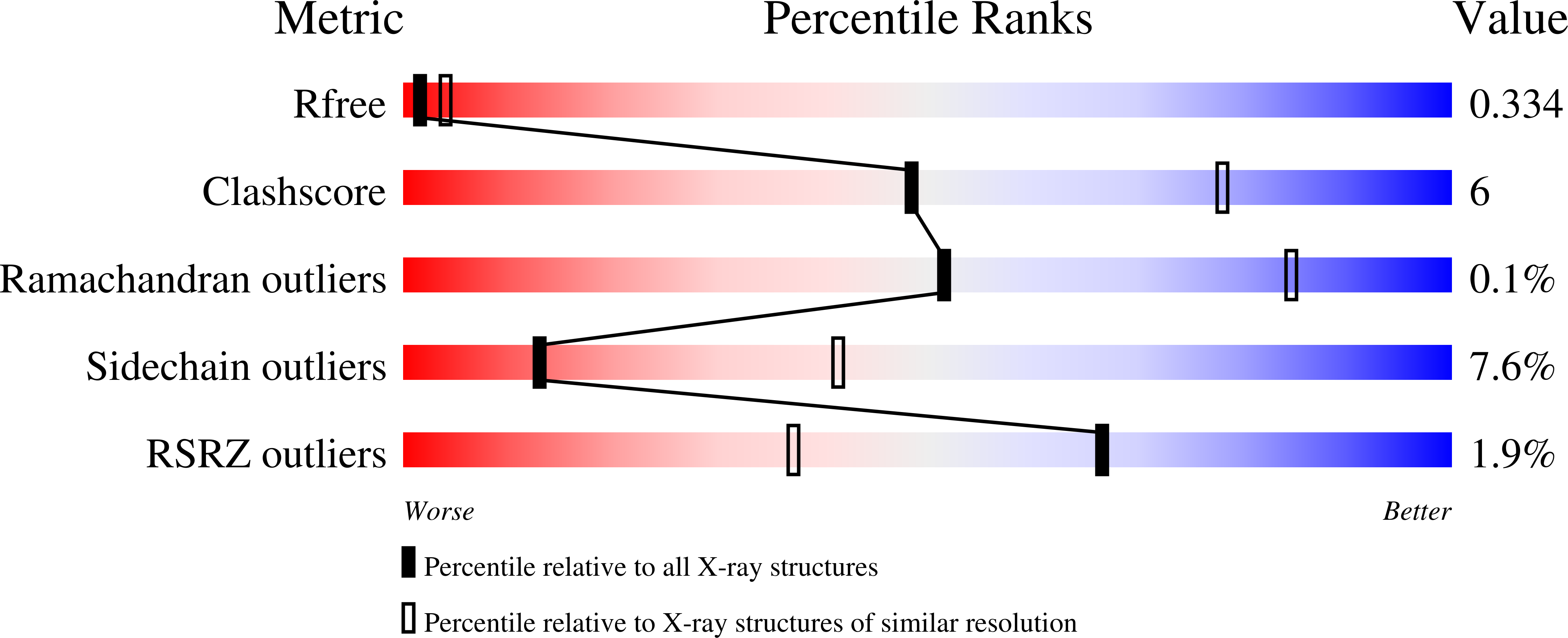
4DCN - PubMed Abstract:
Membrane-sculpting BAR (Bin/Amphiphysin/Rvs) domains form a crescent-shaped homodimer that can sense and induce membrane curvature through its positively charged concave face. We have recently shown that Arfaptin-2, which was originally identified as a binding partner for the Arf and Rac1 GTPases, binds to Arl1 through its BAR domain and is recruited onto Golgi membranes. There, Arfaptin-2 induces membrane tubules. Here, we report the crystal structure of the Arfaptin-2 BAR homodimer in complex with two Arl1 molecules bound symmetrically to each side, leaving the concave face open for membrane association. The overall structure of the Arl1·Arfaptin-2 BAR complex closely resembles that of the PX-BAR domain of sorting nexin 9, suggesting similar mechanisms underlying BAR domain targeting to specific organellar membranes. The Arl1·Arfaptin-2 BAR structure suggests that one of the two Arl1 molecules competes with Rac1, which binds to the concave face of the Arfaptin-2 BAR homodimer and may hinder its membrane association.
Organizational Affiliation:
Structural Biology Research Center, Photon Factory, Institute of Materials Structure Science, High Energy Accelerator Research Organization, KEK, Tsukuba, Ibaraki 305-0801, Japan.