Crystal structure of a predicted N6-adenine-specific DNA methylase from Listeria monocytogenes str. 4b F2365
Nocek, B., Xu, X., Cui, H., Ng, J., Savchenko, A., Edwards, A., Joachimiak, A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| predicted N6-adenine-specific DNA methylase | 393 | Listeria monocytogenes serotype 4b str. F2365 | Mutation(s): 0 Gene Names: LMOf2365_1916 | 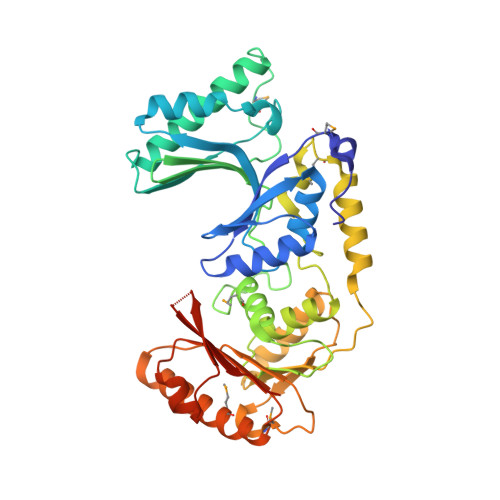 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GOL Query on GOL | C [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | B [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 125.949 | α = 90 |
| b = 46.156 | β = 131.09 |
| c = 91.376 | γ = 90 |
| Software Name | Purpose |
|---|---|
| SBC-Collect | data collection |
| HKL-3000 | phasing |
| SHELX | model building |
| DM | model building |
| ARP/wARP | model building |
| Coot | model building |
| CCP4 | model building |
| MLPHARE | phasing |
| REFMAC | refinement |
| HKL-3000 | data reduction |
| HKL-3000 | data scaling |
| SHELX | phasing |
| DM | phasing |
| CCP4 | phasing |