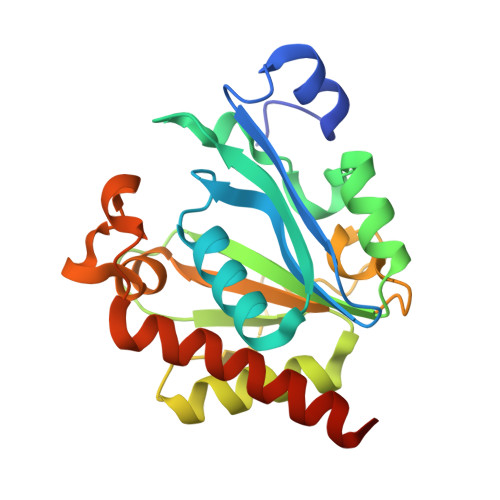
Crystal structure of bacteriophage PhiNIT1 zinc peptidase PghP that hydrolyzes gamma-glutamyl linkage of bacterial poly-gamma-glutamate
Fujimoto, Z., Kimura, K.(2012) Proteins 80: 722-732
- PubMed: 22105902
- DOI: https://doi.org/10.1002/prot.23229
- Primary Citation of Related Structures:
3A9L - PubMed Abstract:
Poly-γ-glutamate hydrolase P (PghP) of Bacillus subtilis bacteriophage ΦNIT1 hydrolyzes the γ-glutamyl peptide linkage of extracellular poly-γ-glutamate produced by bacilli, which facilitates infection and propagation of phage progenies. Crystal structure of PghP was determined at a resolution of 1.9 Å. Structure of PghP was elucidated as a globular protein with an open α/β mixed core structure and a seven-stranded parallel/anti-parallel β-sheet. The β-sheet contained a core four-stranded parallel β-sheet. A zinc-binding motif, His-Glu-His, was identified at the C-terminal end of the β-sheet. Structure analysis demonstrated that PghP, which had not been previously classified into any peptidase/protease family due to lack of amino acid sequence similarity with known enzymes, had a catalytic center containing a zinc ion and an overall topology resembling mammalian carboxypeptidase A and related enzymes. Structural comparisons indicated important amino acid residues of PghP for catalysis and recognition of the γ-peptide bond of poly-γ-glutamate, which was confirmed by site-directed mutagenesis of PghP.
Organizational Affiliation:
Biomolecular Research Unit, National Institute of Agrobiological Sciences, 2-1-2 Kannondai, Tsukuba, Ibaraki 305-8602, Japan. zui@affrc.go.jp