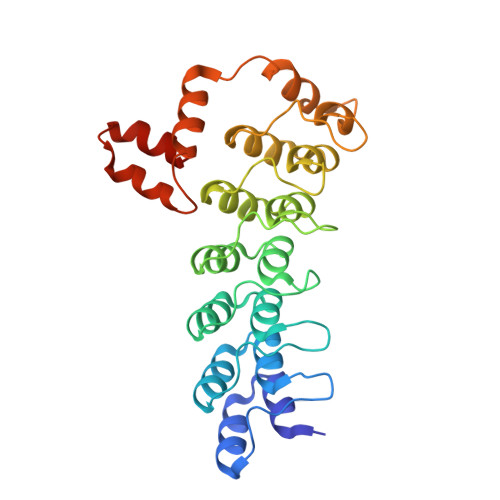
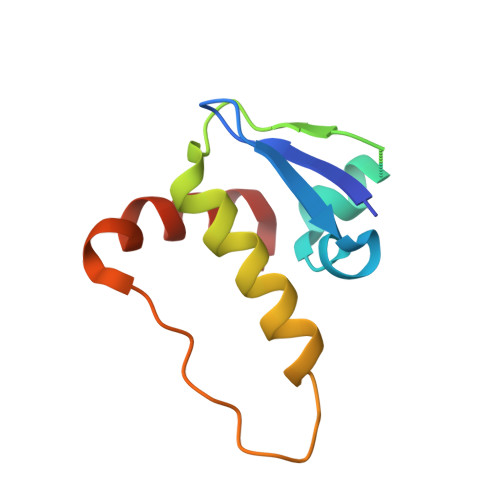
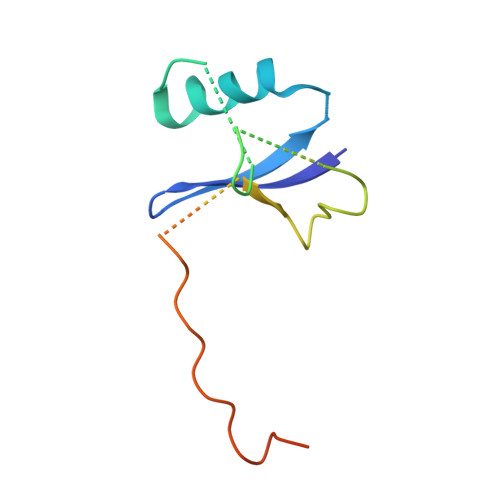
Multimeric Complexes Among Ankyrin-Repeat and Socs-Box Protein 9 (Asb9), Elonginbc, and Cullin 5: Insights Into the Structure and Assembly of Ecs-Type Cullin-Ring E3 Ubiquitin Ligases.
Thomas, J.C., Matak-Vinkovic, D., Van Molle, I., Ciulli, A.(2013) Biochemistry 52: 5236
- PubMed: 23837592
- DOI: https://doi.org/10.1021/bi400758h
- Primary Citation of Related Structures:
3ZNG - PubMed Abstract:
Proteins of the ankyrin-repeat and SOCS-box (ASB) family act as the substrate-recognition subunits of ECS-type (ElonginBC-Cullin-SOCS-box) Cullin RING E3 ubiquitin ligase (CRL) complexes that catalyze the specific polyubiquitination of cellular proteins to target them for degradation by the proteasome. Therefore, ASB multimeric complexes are involved in numerous cell processes and pathways; however, their interactions, assembly, and biological roles remain poorly understood. To enhance our understanding of ASB CRL systems, we investigated the structure, affinity, and assembly of the quaternary multisubunit complex formed by ASB9, Elongin B, Elongin C (EloBC), and Cullin 5. Here, we describe the application of several biophysical techniques including differential scanning fluorimetry, isothermal titration calorimetry (ITC), nanoelectrospray ionization, and ion-mobility mass spectrometry (IM-MS) to provide structural and thermodynamic information for a quaternary ASB CRL complex. We find that ASB9 is unstable alone but forms a stable ternary complex with EloBC that binds with high affinity to the Cullin 5 N-terminal domain (Cul5NTD) but not to Cul2NTD. The structure of the monomeric ASB9-EloBC-Cul5NTD quaternary complex is revealed by molecular modeling and is consistent with IM-MS and temperature-dependent ITC data. This is the first experimental study to validate structural information for the assembly of the quaternary N-terminal region of an ASB CRL complex. The results suggest that ASB E3 ligase complexes function and assemble in an analogous manner to that of other CRL systems and provide a platform for further molecular investigation of this important protein family. The data reported here will also be of use for the future development of chemical probes to examine the biological function and modulation of other ECS-type CRL systems.
Organizational Affiliation:
Department of Chemistry, University of Cambridge , Lensfield Road, Cambridge CB2 1EW, United Kingdom.