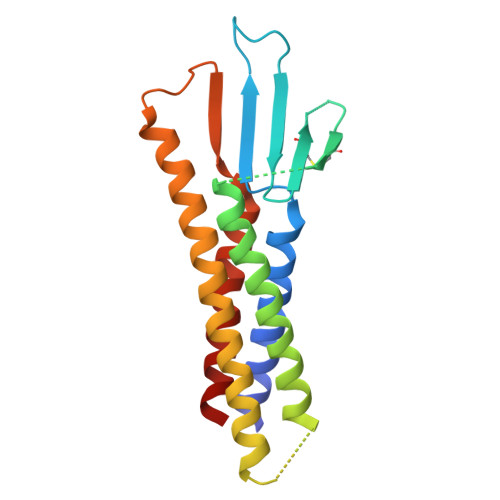
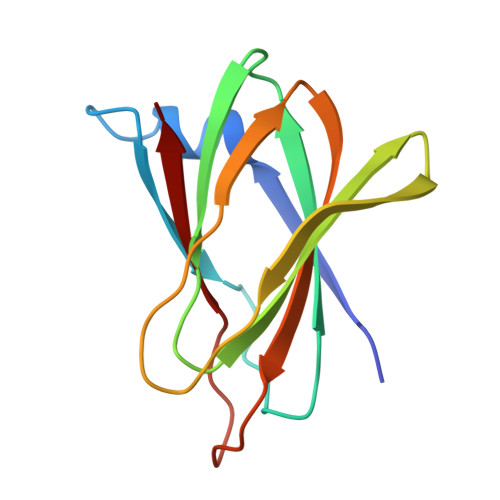
Structural insight into tight junction disassembly by Clostridium perfringens enterotoxin
Saitoh, Y., Suzuki, H., Tani, K., Nishikawa, K., Irie, K., Ogura, Y., Tamura, A., Tsukita, S., Fujiyoshi, Y.(2015) Science 347: 775-778
- PubMed: 25678664
- DOI: https://doi.org/10.1126/science.1261833
- Primary Citation of Related Structures:
3X29 - PubMed Abstract:
The C-terminal region of Clostridium perfringens enterotoxin (C-CPE) can bind to specific claudins, resulting in the disintegration of tight junctions (TJs) and an increase in the paracellular permeability across epithelial cell sheets. Here we present the structure of mammalian claudin-19 in complex with C-CPE at 3.7 Å resolution. The structure shows that C-CPE forms extensive hydrophobic and hydrophilic interactions with the two extracellular segments of claudin-19. The claudin-19/C-CPE complex shows no density of a short extracellular helix that is critical for claudins to assemble into TJ strands. The helix displacement may thus underlie C-CPE-mediated disassembly of TJs.
Organizational Affiliation:
Cellular and Structural Physiology Institute, Nagoya University, Chikusa, Nagoya 464-8601, Japan. Department of Basic Medical Science, Graduate School of Pharmaceutical Science, Nagoya University, Chikusa, Nagoya 464-8601, Japan.