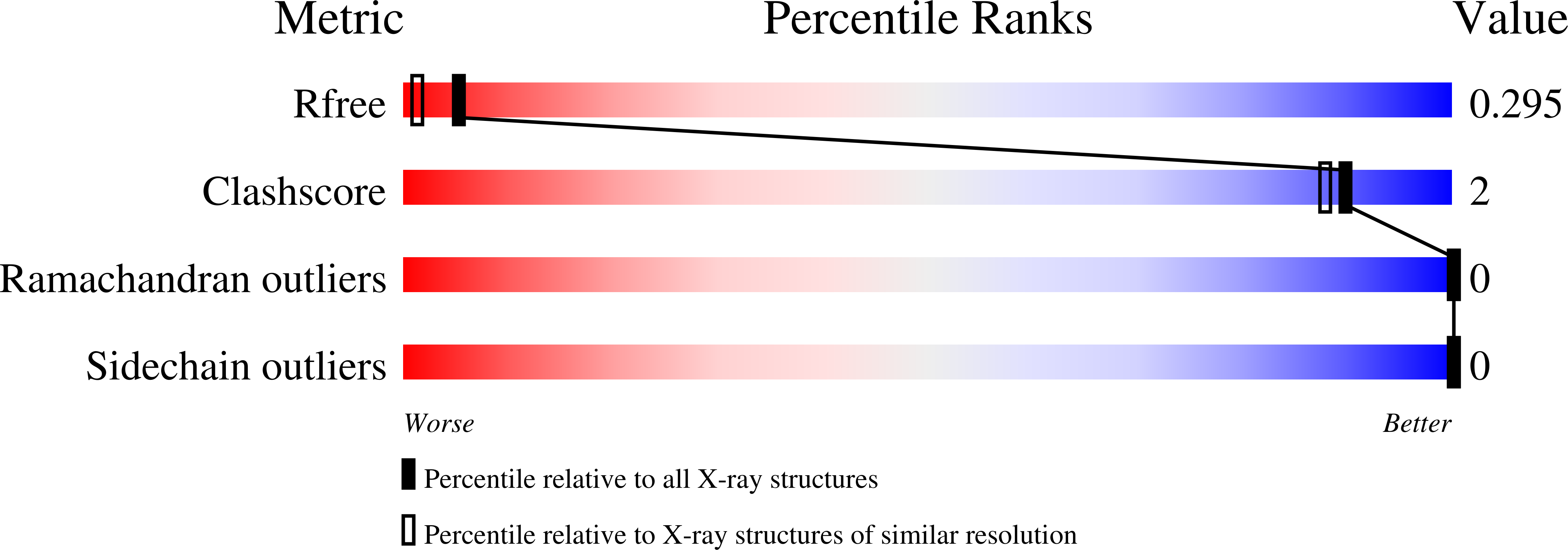Localization and Structure of the Ankyrin-binding Site on beta2-Spectrin
Davis, L., Abdi, K., Machius, M., Brautigam, C., Tomchick, D.R., Bennett, V., Michaely, P.(2009) J Biol Chem 284: 6982-6987
- PubMed: 19098307
- DOI: https://doi.org/10.1074/jbc.M809245200
- Primary Citation of Related Structures:
3EDV - PubMed Abstract:
Spectrins are tetrameric actin-cross-linking proteins that form an elastic network, termed the membrane skeleton, on the cytoplasmic surface of cellular membranes. At the plasma membrane, the membrane skeleton provides essential support, preventing loss of membrane material to environmental shear stresses. The skeleton also controls the location, abundance, and activity of membrane proteins that are critical to cell and tissue function. The ability of the skeleton to modulate membrane stability and function requires adaptor proteins that bind the skeleton to membranes. The principal adaptors are the ankyrin proteins, which bind to the beta-subunit of spectrin and to the cytoplasmic domains of numerous integral membrane proteins. Here, we present the crystal structure of the ankyrin-binding domain of human beta2-spectrin at 1.95 A resolution together with mutagenesis data identifying the binding surface for ankyrins on beta2-spectrin.
Organizational Affiliation:
Department of Cell Biology and Howard Hughes Medical Institute, Duke University Medical Center, Durham, North Carolina 27710, USA.