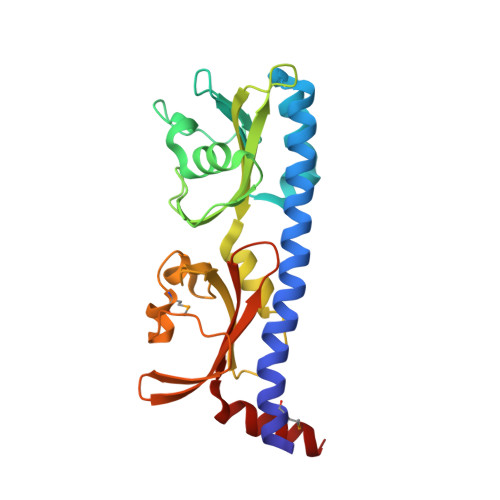
Crystal structure of Mcp_N and cache N-terminal domains of methyl-accepting chemotaxis protein from Vibrio cholerae.
Patskovsky, Y., Ozyurt, S., Freeman, J., Hu, S., Smith, D., Bain, K., Wasserman, S.R., Sauder, J.M., Burley, S.K., Almo, S.C.To be published.