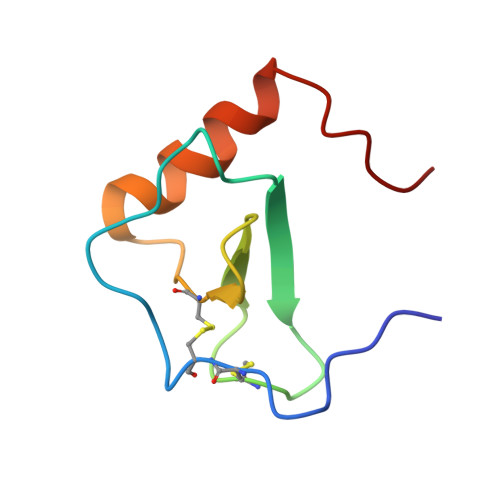
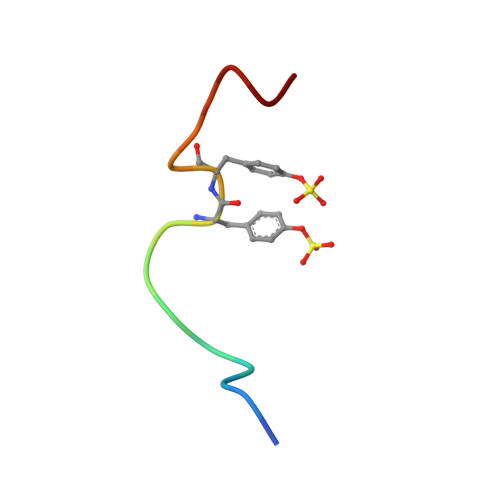
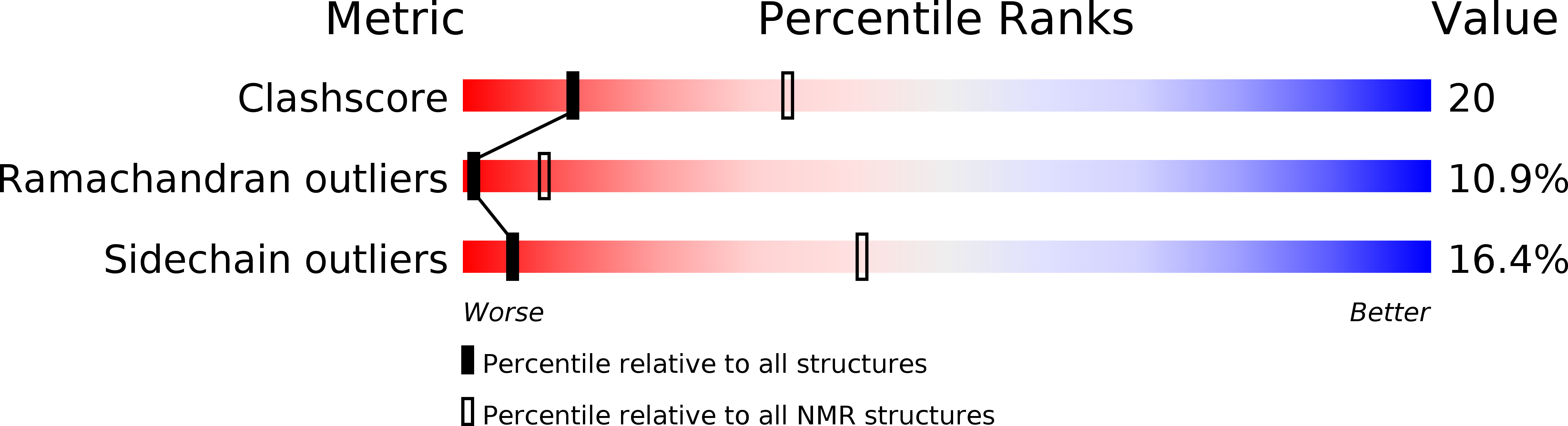Structural Basis of Receptor Sulfotyrosine Recognition by a CC Chemokine: The N-Terminal Region of CCR3 Bound to CCL11/Eotaxin-1.
Millard, C.J., Ludeman, J.P., Canals, M., Bridgford, J.L., Hinds, M.G., Clayton, D.J., Christopoulos, A., Payne, R.J., Stone, M.J.(2014) Structure 22: 1571-1581
- PubMed: 25450766
- DOI: https://doi.org/10.1016/j.str.2014.08.023
- Primary Citation of Related Structures:
2MPM - PubMed Abstract:
Trafficking of leukocytes in immune surveillance and inflammatory responses is activated by chemokines engaging their receptors. Sulfation of tyrosine residues in peptides derived from the eosinophil chemokine receptor CCR3 dramatically enhances binding to cognate chemokines. We report the structural basis of this recognition and affinity enhancement. We describe the structure of a CC chemokine (CCL11/eotaxin-1) bound to a fragment of a chemokine receptor: residues 8–23 of CCR3, including two sulfotyrosine residues. We also show that intact CCR3 is sulfated and sulfation enhances receptor activity. The CCR3 sulfotyrosine residues form hydrophobic, salt bridge and cation-p interactions with residues that are highly conserved in CC chemokines. However, the orientation of the chemokine relative to the receptor N terminus differs substantially from those observed for two CXC chemokines, suggesting that initial binding of the receptor sulfotyrosine residues guides subsequent steps in receptor activation, thereby influencing the receptor conformational changes and signaling.