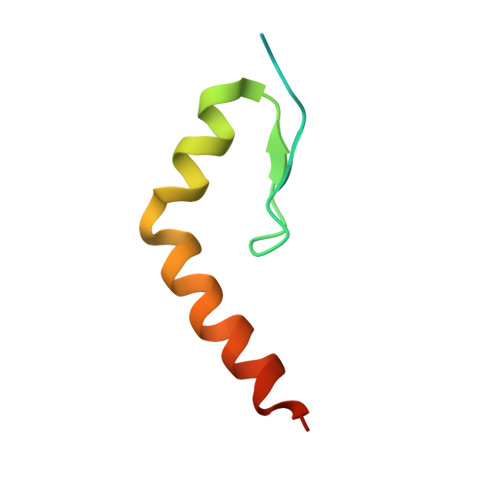
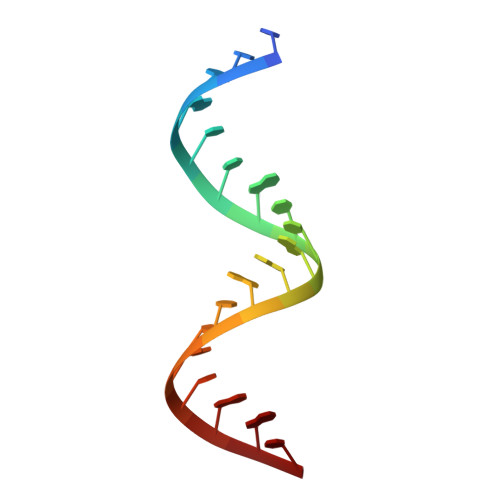
Structural Characterization of Interactions between the Double-Stranded RNA-Binding Zinc Finger Protein JAZ and Nucleic Acids.
Burge, R.G., Martinez-Yamout, M.A., Dyson, H.J., Wright, P.E.(2014) Biochemistry 53: 1495-1510
- PubMed: 24521053
- DOI: https://doi.org/10.1021/bi401675h
- Primary Citation of Related Structures:
2MKD, 2MKN - PubMed Abstract:
The interactions of the human double-stranded RNA-binding zinc finger protein JAZ with RNA or DNA were investigated using electrophoretic mobility-shift assays, isothermal calorimetry, and nuclear magnetic resonance spectroscopy. Consistent with previous reports, JAZ has very low affinity for duplex DNA or single-stranded RNA, but it binds preferentially to double-stranded RNA (dsRNA) with no detectable sequence specificity. The affinity of JAZ for dsRNA is unaffected by local structural features such as loops, overhangs, and bulges, provided a sufficient length of reasonably well-structured A-form RNA (about 18 bp for a single zinc finger) is present. Full-length JAZ contains four Cys2His2 zinc fingers (ZF1-4) and has the highest apparent affinity for dsRNA; two-finger constructs ZF12 and ZF23 have lower affinity, and ZF34 binds even more weakly. The fourth zinc finger, ZF4, has no measurable RNA-binding affinity. Single zinc finger constructs ZF1, ZF2, and ZF3 show evidence for multiple-site binding on the minimal RNA. Fitting of quantitative NMR titration and isothermal calorimetry data to a two-site binding model gave Kd1 ∼ 10 μM and Kd2 ∼ 100 μM. Models of JAZ-RNA complexes were generated using the high-ambiguity-driven biomolecular docking (HADDOCK) program. Single zinc fingers bind to the RNA backbone without sequence specificity, forming complexes with contacts between the RNA minor groove and residues in the N-terminal β strands and between the major groove and residues in the helix-kink-helix motif. We propose that the non-sequence-specific interaction between the zinc fingers of JAZ with dsRNA is dependent only on the overall shape of the A-form RNA.
Organizational Affiliation:
Department of Integrative Structural and Computational Biology and the Skaggs Institute for Chemical Biology, The Scripps Research Institute , La Jolla, California 92037, United States.