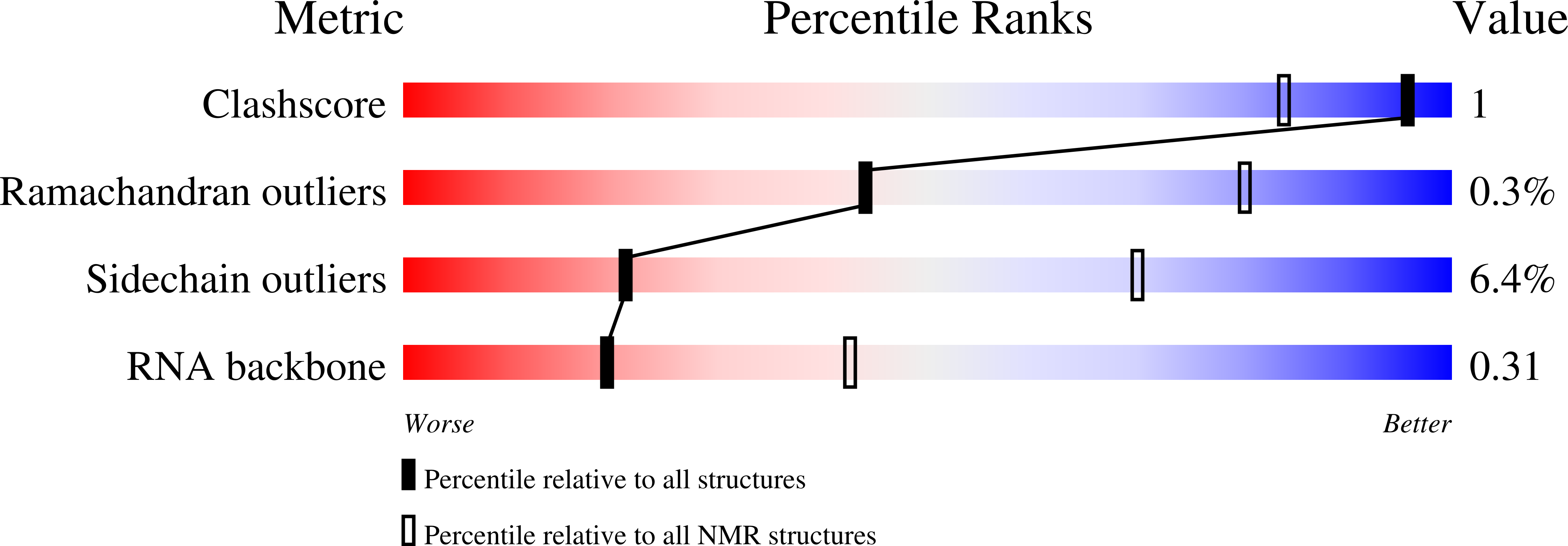
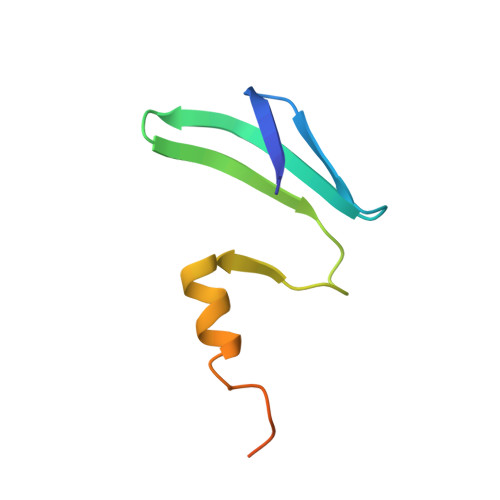
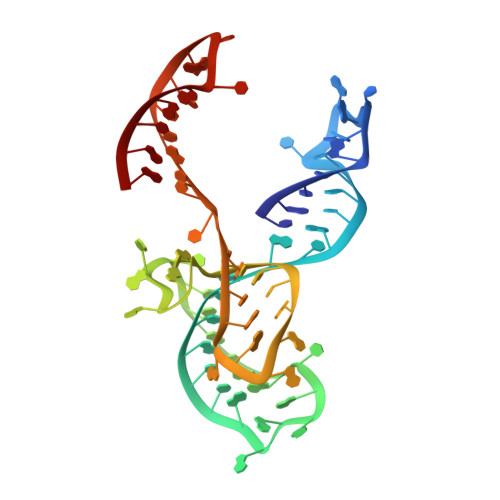
Structural basis of the non-coding RNA RsmZ acting as a protein sponge.
Duss, O., Michel, E., Yulikov, M., Schubert, M., Jeschke, G., Allain, F.H.(2014) Nature 509: 588-592
- PubMed: 24828038
- DOI: https://doi.org/10.1038/nature13271
- Primary Citation of Related Structures:
2MF0, 2MF1 - PubMed Abstract:
MicroRNA and protein sequestration by non-coding RNAs (ncRNAs) has recently generated much interest. In the bacterial Csr/Rsm system, which is considered to be the most general global post-transcriptional regulatory system responsible for bacterial virulence, ncRNAs such as CsrB or RsmZ activate translation initiation by sequestering homodimeric CsrA-type proteins from the ribosome-binding site of a subset of messenger RNAs. However, the mechanism of ncRNA-mediated protein sequestration is not understood at the molecular level. Here we show for Pseudomonas fluorescens that RsmE protein dimers assemble sequentially, specifically and cooperatively onto the ncRNA RsmZ within a narrow affinity range. This assembly yields two different native ribonucleoprotein structures. Using a powerful combination of nuclear magnetic resonance and electron paramagnetic resonance spectroscopy we elucidate these 70-kilodalton solution structures, thereby revealing the molecular mechanism of the sequestration process and how RsmE binding protects the ncRNA from RNase E degradation. Overall, our findings suggest that RsmZ is well-tuned to sequester, store and release RsmE and therefore can be viewed as an ideal protein 'sponge'.
Organizational Affiliation:
Institute of Molecular Biology and Biophysics, ETH Zürich, CH-8093 Zürich, Switzerland.