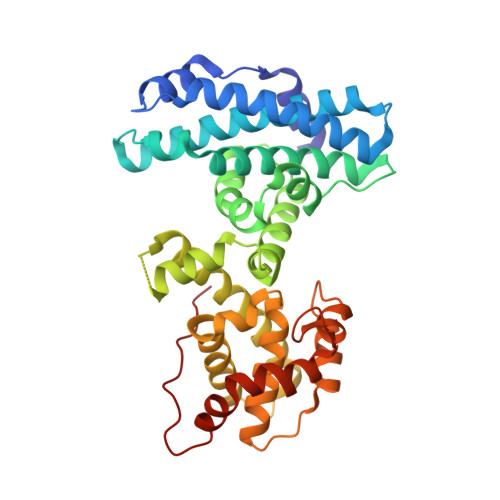
Structure of the retinoblastoma protein bound to adenovirus E1A reveals the molecular basis for viral oncoprotein inactivation of a tumor suppressor
Liu, X., Marmorstein, R.(2007) Genes Dev 21: 2711-2716
- PubMed: 17974914
- DOI: https://doi.org/10.1101/gad.1590607
- Primary Citation of Related Structures:
2R7G - PubMed Abstract:
The adenovirus (Ad) E1A (Ad-E1A) oncoprotein mediates cell transformation, in part, by displacing E2F transcription factors from the retinoblastoma protein (pRb) tumor suppressor. In this study we determined the crystal structure of the pRb pocket domain in complex with conserved region 1 (CR1) of Ad5-E1A. The structure and accompanying biochemical studies reveal that E1A-CR1 binds at the interface of the A and B cyclin folds of the pRb pocket domain, and that both E1A-CR1 and the E2F transactivation domain use similar conserved nonpolar residues to engage overlapping sites on pRb, implicating a novel molecular mechanism for pRb inactivation by a viral oncoprotein.
Organizational Affiliation:
Program in Gene Expression and Regulation, The Wistar Institute, Philadelphia, Pennsylvania 19104, USA.