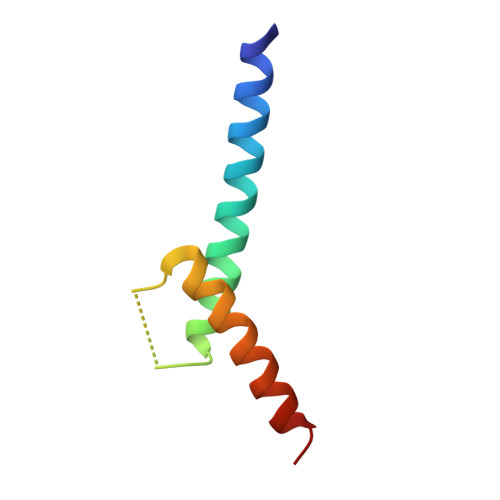
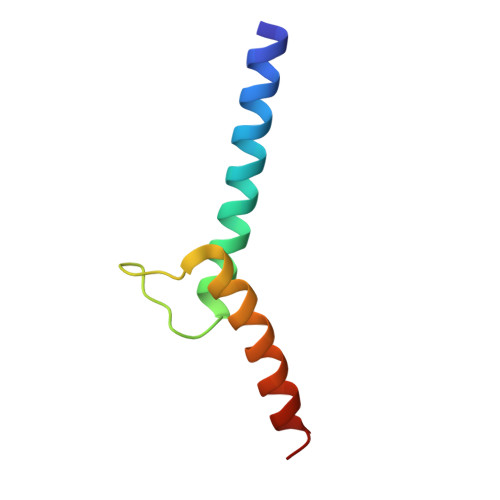
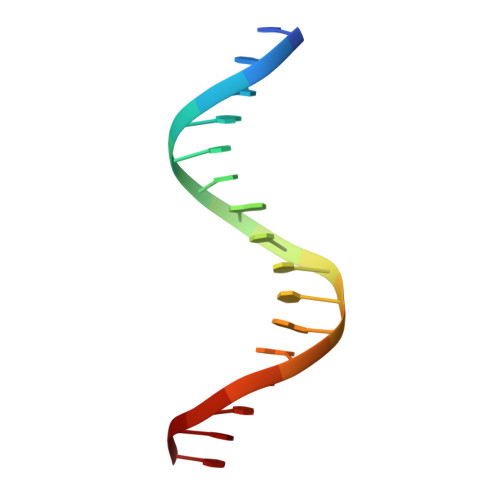
Crystal structure of E47-NeuroD1/beta2 bHLH domain-DNA complex: heterodimer selectivity and DNA recognition.
Longo, A., Guanga, G.P., Rose, R.B.(2008) Biochemistry 47: 218-229
- PubMed: 18069799
- DOI: https://doi.org/10.1021/bi701527r
- Primary Citation of Related Structures:
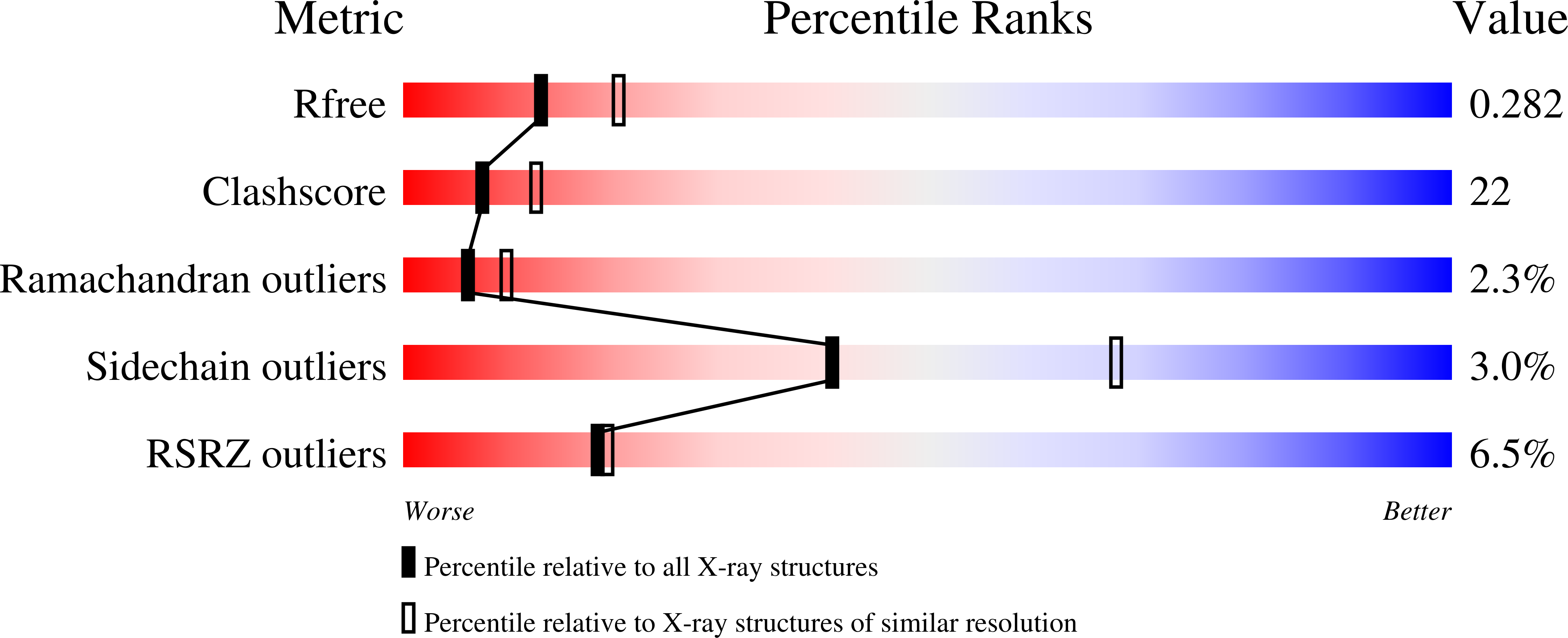
2QL2 - PubMed Abstract:
The ubiquitous class I basic helix-loop-helix (bHLH) factor E47 forms heterodimers with multiple tissue specific class II bHLH proteins to regulate distinct differentiation pathways. In order to define how class I- class II heterodimer partners are selected, we determined the crystal structure of the E47-NeuroD1-bHLH dimer in complex with the insulin promoter E-box sequence. Purification of the bHLH domain of E47-NeuroD1 indicates that E47 heterodimers are stable in solution. The interactions between E47 and NeuroD1 in the heterodimer are comparable to the interactions between E47 monomers in the homodimer, including hydrogen bonding, buried hydrophobic surface, and packing interactions. This is consistent with a model in which E47-NeuroD1 heterodimers are favored due to the instability of NeuroD1 homodimers. Although E47-NeuroD1 is oriented uniquely on the E-box sequence (CATCTG) within the promoter of the insulin gene, no direct contacts are observed with the central base pairs within this E-box sequence. We propose that concerted domain motions allow E47 to form specific base contacts in solution. NeuroD1 is restrained from adopting the same base contacts by an additional phosphate backbone interaction by the neurogenic-specific residue His115. Orienting E47-NeuroD1 on promoters may foster protein-protein contacts essential to initiate transcription.
Organizational Affiliation:
Department of Molecular and Structural Biochemistry, North Carolina State University, 128 Polk Hall, Raleigh, North Carolina 27695-7622, USA.