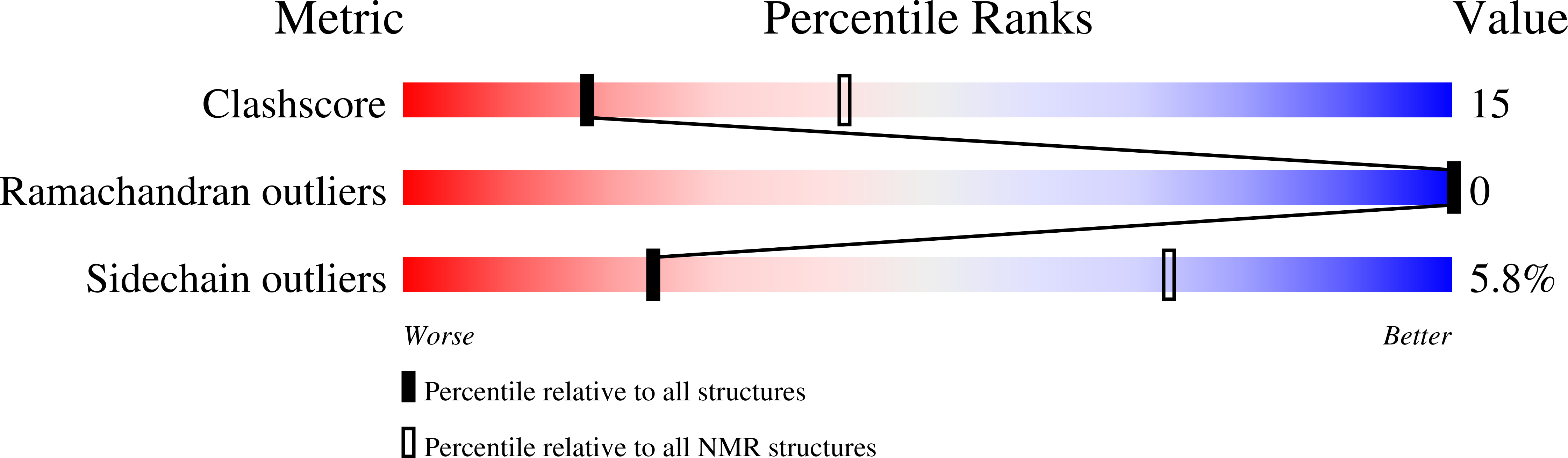A Conserved Ectodomain-Transmembrane Domain Linker Motif Tunes the Allosteric Regulation of Cell Surface Receptors.
Schmidt, T., Ye, F., Situ, A.J., An, W., Ginsberg, M.H., Ulmer, T.S.(2016) J Biol Chem 291: 17536-17546
- PubMed: 27365391
- DOI: https://doi.org/10.1074/jbc.M116.733683
- Primary Citation of Related Structures:
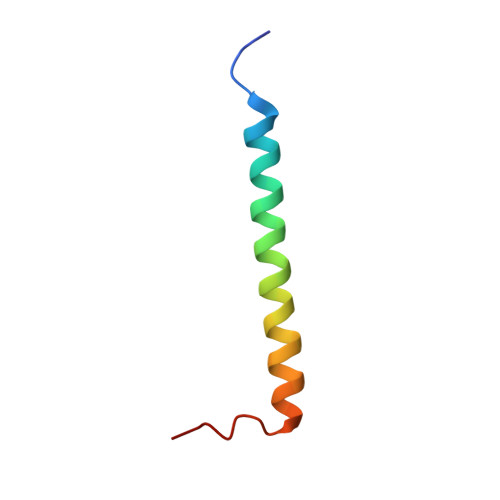
2NA8, 2NA9 - PubMed Abstract:
In many families of cell surface receptors, a single transmembrane (TM) α-helix separates ecto- and cytosolic domains. A defined coupling of ecto- and TM domains must be essential to allosteric receptor regulation but remains little understood. Here, we characterize the linker structure, dynamics, and resulting ecto-TM domain coupling of integrin αIIb in model constructs and relate it to other integrin α subunits by mutagenesis. Cellular integrin activation assays subsequently validate the findings in intact receptors. Our results indicate a flexible yet carefully tuned ecto-TM coupling that modulates the signaling threshold of integrin receptors. Interestingly, a proline at the N-terminal TM helix border, termed NBP, is critical to linker flexibility in integrins. NBP is further predicted in 21% of human single-pass TM proteins and validated in cytokine receptors by the TM domain structure of the cytokine receptor common subunit β and its P441A-substituted variant. Thus, NBP is a conserved uncoupling motif of the ecto-TM domain transition and the degree of ecto-TM domain coupling represents an important parameter in the allosteric regulation of diverse cell surface receptors.
Organizational Affiliation:
From the Department of Biochemistry & Molecular Biology and Zilkha Neurogenetic Institute, Keck School of Medicine, University of Southern California, Los Angeles, California 90033.