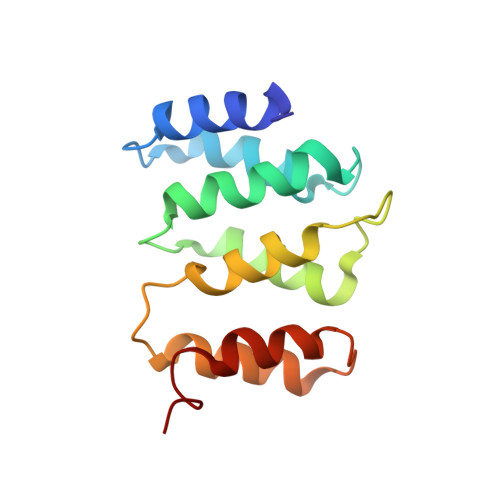
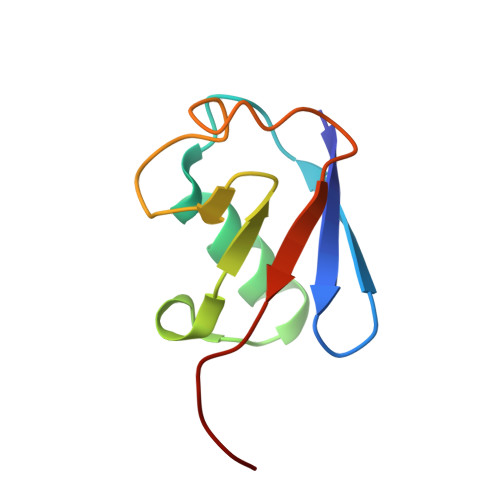
Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome.
Shi, Y., Chen, X., Elsasser, S., Stocks, B.B., Tian, G., Lee, B.H., Shi, Y., Zhang, N., de Poot, S.A., Tuebing, F., Sun, S., Vannoy, J., Tarasov, S.G., Engen, J.R., Finley, D., Walters, K.J.(2016) Science 351
- PubMed: 26912900
- DOI: https://doi.org/10.1126/science.aad9421
- Primary Citation of Related Structures:
2N3T, 2N3U, 2N3V, 2N3W - PubMed Abstract:
Hundreds of pathways for degradation converge at ubiquitin recognition by a proteasome. Here, we found that the five known proteasomal ubiquitin receptors in yeast are collectively nonessential for ubiquitin recognition and identified a sixth receptor, Rpn1. A site ( T1: ) in the Rpn1 toroid recognized ubiquitin and ubiquitin-like ( UBL: ) domains of substrate shuttling factors. T1 structures with monoubiquitin or lysine 48 diubiquitin show three neighboring outer helices engaging two ubiquitins. T1 contributes a distinct substrate-binding pathway with preference for lysine 48-linked chains. Proximal to T1 within the Rpn1 toroid is a second UBL-binding site ( T2: ) that assists in ubiquitin chain disassembly, by binding the UBL of deubiquitinating enzyme Ubp6. Thus, a two-site recognition domain intrinsic to the proteasome uses distinct ubiquitin-fold ligands to assemble substrates, shuttling factors, and a deubiquitinating enzyme.
Organizational Affiliation:
Department of Cell Biology, Harvard Medical School, 240 Longwood Avenue, Boston, MA 02115, USA.